Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
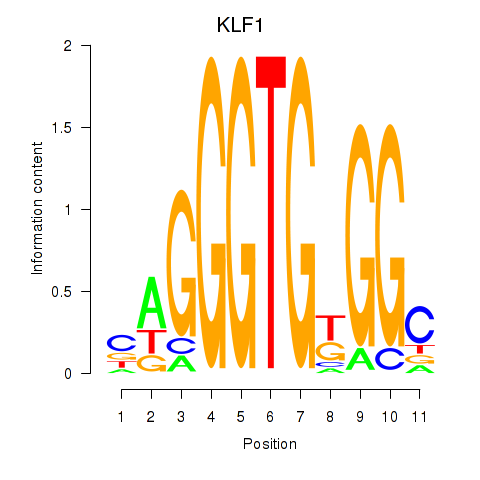
Results for KLF1
Z-value: 1.59
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.6 | KLF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg38_v1_chr19_-_12887188_12887207 | -0.18 | 3.5e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 2.7 | 13.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.7 | 14.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.3 | 3.8 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 1.0 | 3.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 1.0 | 3.0 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.8 | 0.8 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.8 | 4.9 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.8 | 3.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.8 | 9.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.7 | 2.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.7 | 2.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 3.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.6 | 2.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.5 | 2.6 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.5 | 1.5 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.5 | 3.5 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.5 | 2.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.5 | 3.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 | 1.9 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.5 | 1.4 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 0.5 | 9.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.5 | 1.8 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.4 | 5.2 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.4 | 3.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 2.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.4 | 1.6 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.4 | 1.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 5.8 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.4 | 6.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.4 | 2.7 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.3 | 5.7 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.3 | 1.0 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.3 | 4.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 7.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 1.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.3 | 2.5 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 0.9 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.3 | 0.9 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.3 | 1.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 8.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 6.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.3 | 0.8 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.3 | 1.4 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 0.8 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.3 | 1.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.3 | 6.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 1.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 0.7 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.2 | 1.0 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.2 | 1.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 2.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 0.7 | GO:1902688 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) |
| 0.2 | 0.7 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 0.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 3.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.3 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 4.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.2 | 0.7 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.9 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.6 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.2 | 7.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 1.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 1.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 0.8 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.2 | 0.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 0.6 | GO:1903179 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.2 | 24.3 | GO:0070268 | cornification(GO:0070268) |
| 0.2 | 0.5 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.2 | 0.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.5 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.5 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.5 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 1.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 0.6 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.2 | 1.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.2 | 1.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 2.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 5.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.5 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 2.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 2.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 2.2 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 1.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.4 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.1 | 1.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 2.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 3.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 1.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 1.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 1.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 1.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.7 | GO:0060762 | detection of muscle stretch(GO:0035995) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 3.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 5.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.1 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.8 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:0048242 | regulation of epinephrine secretion(GO:0014060) epinephrine secretion(GO:0048242) |
| 0.1 | 0.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 3.7 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.2 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.2 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.6 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 2.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.5 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 1.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.5 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 3.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.7 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 7.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 4.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0033605 | positive regulation of catecholamine secretion(GO:0033605) |
| 0.0 | 1.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 1.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 2.9 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 1.6 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 1.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.6 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 1.6 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 1.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 1.8 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 1.0 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.6 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.8 | GO:0051782 | negative regulation of cell division(GO:0051782) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.8 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.6 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.3 | GO:0060732 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 1.1 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 2.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 1.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 1.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 1.1 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.1 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.0 | 0.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.8 | 13.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.8 | 3.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 2.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.4 | 5.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 7.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 2.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 6.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 6.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 3.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 1.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 7.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.2 | 3.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 2.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 7.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 2.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 3.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 4.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.7 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.2 | 1.4 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 0.5 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.2 | 1.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 4.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.6 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 1.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 6.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 17.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 3.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 3.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 13.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 5.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 4.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 2.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 26.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 7.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.7 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.2 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.5 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.4 | GO:0030395 | lactose binding(GO:0030395) |
| 1.2 | 4.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.0 | 9.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 3.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.8 | 4.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.8 | 3.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.7 | 7.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.7 | 2.7 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.7 | 2.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.7 | 14.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.6 | 1.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.6 | 1.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.5 | 5.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 2.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.5 | 6.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.4 | 1.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.4 | 3.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 6.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 3.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 1.4 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.3 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.9 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 6.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.3 | 3.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 5.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 2.0 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 1.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 2.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 3.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 5.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 1.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 3.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 1.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 3.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.7 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 5.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 2.0 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 3.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 8.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 12.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 3.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 3.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 5.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 2.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 2.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 2.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 7.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 2.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 5.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 2.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 5.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 7.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 3.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 5.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 6.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.7 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 7.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) nerve growth factor binding(GO:0048406) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 7.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 7.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 14.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 14.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 8.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 8.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 3.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 13.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 6.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 5.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 5.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 6.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 4.5 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 1.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 5.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 4.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 3.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 2.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |