Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
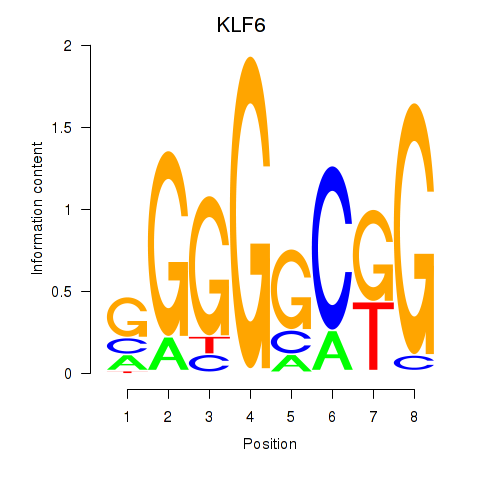
Results for KLF6
Z-value: 0.90
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.15 | KLF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg38_v1_chr10_-_3785197_3785216 | 0.08 | 6.6e-01 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_73874461 | 2.35 |
ENST00000305762.11
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr1_+_7784411 | 2.05 |
ENST00000613533.4
ENST00000614998.4 |
PER3
|
period circadian regulator 3 |
| chr10_+_123135938 | 1.88 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr10_-_73874568 | 1.79 |
ENST00000322635.7
ENST00000322680.7 ENST00000394762.7 ENST00000680035.1 |
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr10_-_73874502 | 1.40 |
ENST00000372765.5
ENST00000351293.7 ENST00000441192.2 ENST00000423381.6 |
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma |
| chr21_+_42219123 | 1.08 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr21_+_42219111 | 1.06 |
ENST00000450121.5
ENST00000361802.6 |
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr19_+_10289939 | 1.00 |
ENST00000221980.5
|
ICAM5
|
intercellular adhesion molecule 5 |
| chr11_-_65872728 | 0.91 |
ENST00000528176.5
ENST00000307998.11 |
EFEMP2
|
EGF containing fibulin extracellular matrix protein 2 |
| chr19_-_45496998 | 0.80 |
ENST00000245923.9
ENST00000590526.5 ENST00000344680.8 |
RTN2
|
reticulon 2 |
| chr6_-_169723931 | 0.75 |
ENST00000366780.8
ENST00000612128.1 |
PHF10
|
PHD finger protein 10 |
| chr19_+_45001430 | 0.72 |
ENST00000625761.2
ENST00000505236.1 ENST00000221452.13 |
RELB
|
RELB proto-oncogene, NF-kB subunit |
| chr11_-_2885728 | 0.65 |
ENST00000647251.1
ENST00000380725.2 ENST00000430149.3 ENST00000414822.8 ENST00000440480.8 |
CDKN1C
|
cyclin dependent kinase inhibitor 1C |
| chr16_+_31459479 | 0.65 |
ENST00000268314.9
|
ARMC5
|
armadillo repeat containing 5 |
| chr2_-_25252072 | 0.64 |
ENST00000683760.1
|
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr6_-_29628038 | 0.64 |
ENST00000355973.7
ENST00000377012.8 |
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr7_-_100177336 | 0.63 |
ENST00000292377.4
|
GPC2
|
glypican 2 |
| chr1_-_221742074 | 0.62 |
ENST00000366899.4
|
DUSP10
|
dual specificity phosphatase 10 |
| chr11_-_64166102 | 0.62 |
ENST00000255681.7
ENST00000675777.1 |
MACROD1
|
mono-ADP ribosylhydrolase 1 |
| chr16_-_1782526 | 0.61 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr1_+_6613722 | 0.61 |
ENST00000377648.5
|
PHF13
|
PHD finger protein 13 |
| chr16_+_31459950 | 0.61 |
ENST00000564900.1
|
ARMC5
|
armadillo repeat containing 5 |
| chr19_+_45497246 | 0.59 |
ENST00000396737.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr1_+_37474572 | 0.59 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr19_+_45497221 | 0.59 |
ENST00000456399.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr14_+_104724221 | 0.58 |
ENST00000330877.7
|
ADSS1
|
adenylosuccinate synthase 1 |
| chr17_+_4807119 | 0.57 |
ENST00000263088.11
ENST00000572940.5 |
PLD2
|
phospholipase D2 |
| chr1_+_156860886 | 0.57 |
ENST00000358660.3
ENST00000675461.1 |
NTRK1
|
neurotrophic receptor tyrosine kinase 1 |
| chr1_+_156860857 | 0.57 |
ENST00000524377.7
|
NTRK1
|
neurotrophic receptor tyrosine kinase 1 |
| chr16_+_74999312 | 0.57 |
ENST00000566250.5
ENST00000567962.5 |
ZNRF1
|
zinc and ring finger 1 |
| chr1_+_156860815 | 0.56 |
ENST00000368196.7
|
NTRK1
|
neurotrophic receptor tyrosine kinase 1 |
| chr7_-_143362687 | 0.56 |
ENST00000409578.5
ENST00000443739.7 ENST00000409346.5 |
FAM131B
|
family with sequence similarity 131 member B |
| chr6_-_31729785 | 0.54 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_-_25252251 | 0.54 |
ENST00000380746.8
ENST00000402667.1 |
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr17_+_45221993 | 0.53 |
ENST00000328118.7
|
FMNL1
|
formin like 1 |
| chr11_-_46918522 | 0.53 |
ENST00000378623.6
ENST00000534404.1 |
LRP4
|
LDL receptor related protein 4 |
| chr3_-_125055987 | 0.52 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr11_+_118607579 | 0.51 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr14_+_94174284 | 0.51 |
ENST00000304338.8
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr11_-_560702 | 0.50 |
ENST00000441853.5
ENST00000329451.8 |
LMNTD2
|
lamin tail domain containing 2 |
| chr1_+_7784251 | 0.49 |
ENST00000377532.8
ENST00000377541.5 |
PER3
|
period circadian regulator 3 |
| chr2_-_85888958 | 0.49 |
ENST00000377332.8
ENST00000639119.1 ENST00000638572.2 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr3_+_49171590 | 0.48 |
ENST00000332780.4
|
KLHDC8B
|
kelch domain containing 8B |
| chr22_-_38088915 | 0.48 |
ENST00000428572.1
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr1_+_156728442 | 0.48 |
ENST00000368218.8
ENST00000368216.9 |
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr5_+_93584916 | 0.48 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr13_+_31846713 | 0.47 |
ENST00000645780.1
|
FRY
|
FRY microtubule binding protein |
| chr14_+_94174334 | 0.46 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr10_+_97713694 | 0.46 |
ENST00000285605.8
|
MARVELD1
|
MARVEL domain containing 1 |
| chr19_+_48170654 | 0.46 |
ENST00000614654.2
ENST00000328759.11 |
ZSWIM9
|
zinc finger SWIM-type containing 9 |
| chr2_-_85888685 | 0.46 |
ENST00000638178.1
ENST00000640982.1 ENST00000640992.1 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr2_-_25673511 | 0.45 |
ENST00000407186.5
ENST00000404103.7 ENST00000405222.5 ENST00000406818.7 ENST00000407038.7 ENST00000407661.7 ENST00000288642.12 |
DTNB
|
dystrobrevin beta |
| chr9_+_35605234 | 0.45 |
ENST00000336395.6
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr4_-_2262082 | 0.45 |
ENST00000337190.7
|
MXD4
|
MAX dimerization protein 4 |
| chr19_-_1568301 | 0.45 |
ENST00000402693.5
|
MEX3D
|
mex-3 RNA binding family member D |
| chr2_+_69915041 | 0.45 |
ENST00000540449.5
|
MXD1
|
MAX dimerization protein 1 |
| chr7_-_27130733 | 0.45 |
ENST00000428284.2
ENST00000360046.10 ENST00000610970.1 |
HOXA4
|
homeobox A4 |
| chr17_+_4833331 | 0.44 |
ENST00000355280.11
ENST00000347992.11 |
MINK1
|
misshapen like kinase 1 |
| chr9_+_35605277 | 0.44 |
ENST00000620767.4
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr11_+_63813384 | 0.44 |
ENST00000294244.9
|
SPINDOC
|
spindlin interactor and repressor of chromatin binding |
| chr16_-_56425424 | 0.43 |
ENST00000290649.10
|
AMFR
|
autocrine motility factor receptor |
| chr6_-_93419545 | 0.43 |
ENST00000369297.1
ENST00000369303.9 ENST00000680224.1 ENST00000681532.1 ENST00000679565.1 |
EPHA7
|
EPH receptor A7 |
| chr19_-_48511793 | 0.43 |
ENST00000600059.6
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr1_-_23424692 | 0.43 |
ENST00000374601.7
ENST00000450454.7 |
TCEA3
|
transcription elongation factor A3 |
| chr11_+_64234569 | 0.41 |
ENST00000309422.7
ENST00000426086.3 |
VEGFB
|
vascular endothelial growth factor B |
| chr2_+_219442023 | 0.41 |
ENST00000431523.5
ENST00000396698.5 |
SPEG
|
striated muscle enriched protein kinase |
| chr2_+_148021001 | 0.41 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr1_+_151282262 | 0.41 |
ENST00000336715.11
ENST00000324048.9 |
ZNF687
|
zinc finger protein 687 |
| chr1_-_21937300 | 0.40 |
ENST00000374695.8
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr2_-_85888897 | 0.40 |
ENST00000639305.1
ENST00000638986.1 |
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr11_+_57597563 | 0.40 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr5_+_136028979 | 0.40 |
ENST00000442011.7
|
TGFBI
|
transforming growth factor beta induced |
| chr1_-_44031446 | 0.39 |
ENST00000372310.8
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr2_+_148021083 | 0.39 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr19_+_55339867 | 0.39 |
ENST00000255613.8
|
KMT5C
|
lysine methyltransferase 5C |
| chr3_+_50155305 | 0.39 |
ENST00000002829.8
ENST00000426511.5 |
SEMA3F
|
semaphorin 3F |
| chr16_-_67968553 | 0.38 |
ENST00000576616.5
ENST00000572037.5 ENST00000316341.8 ENST00000422611.6 |
SLC12A4
|
solute carrier family 12 member 4 |
| chr18_-_5295679 | 0.38 |
ENST00000615385.4
ENST00000582388.5 ENST00000651870.1 |
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr9_+_125747345 | 0.38 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr9_-_107489754 | 0.38 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr3_+_50155024 | 0.38 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr16_-_30123203 | 0.38 |
ENST00000395202.5
ENST00000395199.7 ENST00000263025.9 ENST00000322266.9 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr11_-_64744317 | 0.38 |
ENST00000419843.1
ENST00000394430.5 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr20_+_36573589 | 0.37 |
ENST00000373872.9
ENST00000650844.1 |
TGIF2
|
TGFB induced factor homeobox 2 |
| chr9_-_91423819 | 0.37 |
ENST00000297689.4
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr11_+_57598184 | 0.37 |
ENST00000677625.1
ENST00000676670.1 |
SERPING1
|
serpin family G member 1 |
| chr15_+_97960692 | 0.36 |
ENST00000268042.7
|
ARRDC4
|
arrestin domain containing 4 |
| chr17_-_7217206 | 0.36 |
ENST00000447163.6
ENST00000647975.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr6_+_37170133 | 0.36 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr20_+_36573458 | 0.36 |
ENST00000373874.6
|
TGIF2
|
TGFB induced factor homeobox 2 |
| chr5_+_79236092 | 0.36 |
ENST00000396137.5
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr15_-_75451543 | 0.36 |
ENST00000394949.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chrX_-_129654946 | 0.35 |
ENST00000429967.3
|
APLN
|
apelin |
| chr6_-_110179995 | 0.35 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr2_+_69915100 | 0.35 |
ENST00000264444.7
|
MXD1
|
MAX dimerization protein 1 |
| chr9_-_37465402 | 0.35 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr10_-_43267059 | 0.35 |
ENST00000395810.6
|
RASGEF1A
|
RasGEF domain family member 1A |
| chr19_+_49581304 | 0.35 |
ENST00000246794.10
|
PRRG2
|
proline rich and Gla domain 2 |
| chr5_+_38846002 | 0.34 |
ENST00000274276.8
|
OSMR
|
oncostatin M receptor |
| chr6_-_31958935 | 0.34 |
ENST00000441998.5
ENST00000444811.6 ENST00000375429.8 |
NELFE
|
negative elongation factor complex member E |
| chr19_+_41354145 | 0.34 |
ENST00000604123.5
|
TMEM91
|
transmembrane protein 91 |
| chr10_+_97498881 | 0.34 |
ENST00000370664.4
|
UBTD1
|
ubiquitin domain containing 1 |
| chr2_-_96116565 | 0.34 |
ENST00000620793.2
|
ADRA2B
|
adrenoceptor alpha 2B |
| chr22_+_41381923 | 0.34 |
ENST00000266304.9
|
TEF
|
TEF transcription factor, PAR bZIP family member |
| chr5_+_38845824 | 0.33 |
ENST00000502536.5
|
OSMR
|
oncostatin M receptor |
| chr1_-_113812254 | 0.33 |
ENST00000616024.4
ENST00000615321.1 |
RSBN1
|
round spermatid basic protein 1 |
| chr8_-_100309904 | 0.33 |
ENST00000523481.5
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr5_-_135535955 | 0.33 |
ENST00000314744.6
|
NEUROG1
|
neurogenin 1 |
| chr6_-_31958852 | 0.33 |
ENST00000375425.9
ENST00000426722.5 |
NELFE
|
negative elongation factor complex member E |
| chr6_+_21593742 | 0.33 |
ENST00000244745.4
|
SOX4
|
SRY-box transcription factor 4 |
| chr6_-_170291053 | 0.33 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr11_+_119168705 | 0.33 |
ENST00000409109.6
ENST00000409991.5 ENST00000292199.6 |
NLRX1
|
NLR family member X1 |
| chr6_-_159727324 | 0.33 |
ENST00000401980.3
ENST00000545162.5 |
SOD2
|
superoxide dismutase 2 |
| chr7_-_143408848 | 0.33 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr17_-_7217178 | 0.32 |
ENST00000485100.5
|
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr6_+_82364234 | 0.32 |
ENST00000543496.3
|
TPBG
|
trophoblast glycoprotein |
| chr11_-_12009082 | 0.32 |
ENST00000396505.7
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_+_210329294 | 0.32 |
ENST00000367010.5
|
HHAT
|
hedgehog acyltransferase |
| chr4_-_36244438 | 0.32 |
ENST00000303965.9
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_+_119168188 | 0.32 |
ENST00000454811.5
ENST00000409265.8 ENST00000449394.5 |
NLRX1
|
NLR family member X1 |
| chr20_-_31870216 | 0.32 |
ENST00000486996.5
ENST00000398084.6 |
DUSP15
|
dual specificity phosphatase 15 |
| chr16_-_57536543 | 0.32 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr16_-_30122944 | 0.32 |
ENST00000484663.5
ENST00000478356.5 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr11_-_1622093 | 0.32 |
ENST00000616115.1
ENST00000399682.1 |
KRTAP5-4
|
keratin associated protein 5-4 |
| chr5_-_148654522 | 0.31 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr16_+_68645290 | 0.31 |
ENST00000264012.9
|
CDH3
|
cadherin 3 |
| chr11_+_68460712 | 0.31 |
ENST00000528635.5
ENST00000533127.5 ENST00000529907.5 ENST00000529344.5 ENST00000534534.5 ENST00000524845.5 ENST00000393800.7 ENST00000265637.8 ENST00000524904.5 ENST00000393801.7 ENST00000265636.9 ENST00000529710.5 |
PPP6R3
|
protein phosphatase 6 regulatory subunit 3 |
| chr17_+_72121012 | 0.31 |
ENST00000245479.3
|
SOX9
|
SRY-box transcription factor 9 |
| chr1_-_44031352 | 0.31 |
ENST00000372306.7
ENST00000475075.6 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr17_-_44363839 | 0.31 |
ENST00000293443.12
|
FAM171A2
|
family with sequence similarity 171 member A2 |
| chr20_-_57710001 | 0.31 |
ENST00000341744.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_110179702 | 0.30 |
ENST00000392587.6
|
WASF1
|
WASP family member 1 |
| chr1_-_35557378 | 0.30 |
ENST00000325722.8
ENST00000469892.5 |
KIAA0319L
|
KIAA0319 like |
| chr7_-_99144053 | 0.30 |
ENST00000361125.1
ENST00000361368.7 |
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr11_+_111602380 | 0.30 |
ENST00000304987.4
|
SIK2
|
salt inducible kinase 2 |
| chr8_+_26383043 | 0.30 |
ENST00000380629.7
|
BNIP3L
|
BCL2 interacting protein 3 like |
| chr17_-_181640 | 0.30 |
ENST00000613549.3
|
DOC2B
|
double C2 domain beta |
| chr15_-_60592507 | 0.29 |
ENST00000449337.6
|
RORA
|
RAR related orphan receptor A |
| chr1_-_32362081 | 0.29 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229 member A |
| chr17_+_76736530 | 0.29 |
ENST00000621483.4
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr6_-_44265541 | 0.28 |
ENST00000619360.6
|
NFKBIE
|
NFKB inhibitor epsilon |
| chr11_+_62613236 | 0.28 |
ENST00000278833.4
|
ROM1
|
retinal outer segment membrane protein 1 |
| chr19_-_1401487 | 0.28 |
ENST00000640762.1
ENST00000252288.8 ENST00000447102.8 |
GAMT
|
guanidinoacetate N-methyltransferase |
| chr19_+_42284020 | 0.28 |
ENST00000160740.7
|
CIC
|
capicua transcriptional repressor |
| chr19_+_35000426 | 0.28 |
ENST00000411896.6
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr11_+_94973640 | 0.28 |
ENST00000335080.6
ENST00000536741.1 |
KDM4D
|
lysine demethylase 4D |
| chr2_+_48440757 | 0.28 |
ENST00000294952.13
ENST00000281394.8 ENST00000449090.6 |
PPP1R21
|
protein phosphatase 1 regulatory subunit 21 |
| chr2_+_30146941 | 0.28 |
ENST00000379520.7
ENST00000379519.7 |
YPEL5
|
yippee like 5 |
| chr12_-_94650506 | 0.27 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr9_+_34958254 | 0.27 |
ENST00000242315.3
|
PHF24
|
PHD finger protein 24 |
| chr6_+_31959130 | 0.27 |
ENST00000375394.7
ENST00000628157.1 |
SKIV2L
|
Ski2 like RNA helicase |
| chr8_-_22232020 | 0.27 |
ENST00000454243.7
ENST00000321613.7 |
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr1_+_226223618 | 0.27 |
ENST00000542034.5
ENST00000366810.6 |
MIXL1
|
Mix paired-like homeobox |
| chr13_+_97434154 | 0.27 |
ENST00000245304.5
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chrX_+_151694967 | 0.26 |
ENST00000448726.5
ENST00000538575.5 |
PRRG3
|
proline rich and Gla domain 3 |
| chr3_+_124584625 | 0.26 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr19_+_531750 | 0.26 |
ENST00000215574.9
|
CDC34
|
cell division cycle 34, ubiqiutin conjugating enzyme |
| chr7_-_140478926 | 0.26 |
ENST00000480552.5
|
MKRN1
|
makorin ring finger protein 1 |
| chr11_-_45665578 | 0.26 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1 |
| chr22_+_37608826 | 0.26 |
ENST00000405147.7
ENST00000343632.9 ENST00000429218.5 ENST00000325180.12 |
GGA1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr20_+_58891704 | 0.26 |
ENST00000371081.5
ENST00000338783.7 |
GNAS
|
GNAS complex locus |
| chr5_-_126595237 | 0.26 |
ENST00000637206.1
ENST00000553117.5 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr11_+_118607598 | 0.26 |
ENST00000600882.6
ENST00000356063.9 |
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr16_+_68644988 | 0.26 |
ENST00000429102.6
|
CDH3
|
cadherin 3 |
| chr9_+_125748175 | 0.26 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr2_+_46941199 | 0.26 |
ENST00000319190.11
ENST00000394850.6 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr5_+_109689915 | 0.26 |
ENST00000261483.5
|
MAN2A1
|
mannosidase alpha class 2A member 1 |
| chr5_-_126595185 | 0.26 |
ENST00000510111.6
ENST00000635851.1 ENST00000637964.1 ENST00000413020.6 ENST00000637782.1 ENST00000409134.8 ENST00000637272.1 ENST00000636879.1 ENST00000636743.1 ENST00000636886.1 ENST00000509270.2 |
ALDH7A1
|
aldehyde dehydrogenase 7 family member A1 |
| chr11_-_63768762 | 0.25 |
ENST00000433688.2
|
C11orf95
|
chromosome 11 open reading frame 95 |
| chr12_+_53380141 | 0.25 |
ENST00000551969.5
ENST00000327443.9 |
SP1
|
Sp1 transcription factor |
| chr5_-_159209503 | 0.25 |
ENST00000424310.7
ENST00000611185.4 |
RNF145
|
ring finger protein 145 |
| chr11_-_417304 | 0.25 |
ENST00000397632.7
|
SIGIRR
|
single Ig and TIR domain containing |
| chr1_-_114153863 | 0.25 |
ENST00000610222.3
ENST00000369547.6 ENST00000641643.2 |
SYT6
|
synaptotagmin 6 |
| chr19_+_47256518 | 0.25 |
ENST00000643617.1
ENST00000221922.11 |
CCDC9
|
coiled-coil domain containing 9 |
| chr15_-_70096604 | 0.25 |
ENST00000559048.5
ENST00000560939.5 ENST00000440567.7 ENST00000557907.5 ENST00000558379.5 ENST00000559929.5 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr2_-_218568291 | 0.25 |
ENST00000418019.5
ENST00000454775.5 ENST00000338465.5 ENST00000415516.5 ENST00000258399.8 |
USP37
|
ubiquitin specific peptidase 37 |
| chr17_-_2025289 | 0.25 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chr6_+_72622086 | 0.25 |
ENST00000370392.5
ENST00000629977.2 |
KCNQ5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr17_+_63477052 | 0.24 |
ENST00000290866.10
ENST00000428043.5 |
ACE
|
angiotensin I converting enzyme |
| chr1_-_20486197 | 0.24 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr19_+_4153616 | 0.24 |
ENST00000078445.7
ENST00000595923.5 ENST00000602257.5 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3 like 3 |
| chr20_-_31870510 | 0.24 |
ENST00000339738.10
|
DUSP15
|
dual specificity phosphatase 15 |
| chr17_-_21253398 | 0.24 |
ENST00000611551.1
|
NATD1
|
N-acetyltransferase domain containing 1 |
| chr19_-_4124082 | 0.24 |
ENST00000394867.8
ENST00000262948.10 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr15_-_55743086 | 0.24 |
ENST00000561292.1
ENST00000389286.9 |
PRTG
|
protogenin |
| chr2_+_219418369 | 0.24 |
ENST00000373960.4
|
DES
|
desmin |
| chr3_-_123449083 | 0.24 |
ENST00000462833.6
|
ADCY5
|
adenylate cyclase 5 |
| chrX_+_54808359 | 0.24 |
ENST00000375058.5
ENST00000375060.5 |
MAGED2
|
MAGE family member D2 |
| chr7_-_27130182 | 0.24 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr18_+_62523002 | 0.24 |
ENST00000269499.10
|
ZCCHC2
|
zinc finger CCHC-type containing 2 |
| chr9_+_126914844 | 0.24 |
ENST00000373436.5
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_-_113812448 | 0.24 |
ENST00000612242.4
ENST00000261441.9 |
RSBN1
|
round spermatid basic protein 1 |
| chr14_-_99480784 | 0.24 |
ENST00000329331.7
ENST00000436070.6 |
SETD3
|
SET domain containing 3, actin histidine methyltransferase |
| chr10_+_100997040 | 0.24 |
ENST00000370223.7
|
LZTS2
|
leucine zipper tumor suppressor 2 |
| chr11_-_72642450 | 0.23 |
ENST00000444035.6
ENST00000544570.5 |
PDE2A
|
phosphodiesterase 2A |
| chr1_+_32886456 | 0.23 |
ENST00000373467.4
|
HPCA
|
hippocalcin |
| chr10_+_87863595 | 0.23 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog |
| chrX_+_48508949 | 0.23 |
ENST00000359882.8
ENST00000326194.11 ENST00000355961.8 ENST00000683923.1 ENST00000489940.5 ENST00000361988.7 |
PORCN
|
porcupine O-acyltransferase |
| chr17_-_63700100 | 0.23 |
ENST00000578993.5
ENST00000259006.8 ENST00000583211.5 |
LIMD2
|
LIM domain containing 2 |
| chr7_+_92245960 | 0.23 |
ENST00000265742.8
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.3 | 1.7 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.3 | 0.8 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 5.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 0.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 0.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 1.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 0.6 | GO:1902908 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.2 | 1.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 0.5 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 0.6 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.2 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.5 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.9 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.5 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.4 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) negative regulation by host of viral genome replication(GO:0044828) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.3 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.4 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.8 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.4 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.1 | 1.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.2 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.2 | GO:0034970 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.1 | 2.7 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.4 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.3 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.3 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.2 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.2 | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.1 | 0.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.2 | GO:1903984 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.4 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.2 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0046668 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) regulation of retinal cell programmed cell death(GO:0046668) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.2 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.4 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.0 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.7 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.3 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.2 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1900163 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 0.0 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.3 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 2.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0032525 | negative regulation of neuron maturation(GO:0014043) somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.2 | GO:0048672 | regulation of collateral sprouting(GO:0048670) positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.4 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.4 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 5.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 5.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.3 | 1.3 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 1.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.8 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0031711 | peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0097001 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 2.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |