Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for LEF1
Z-value: 1.06
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
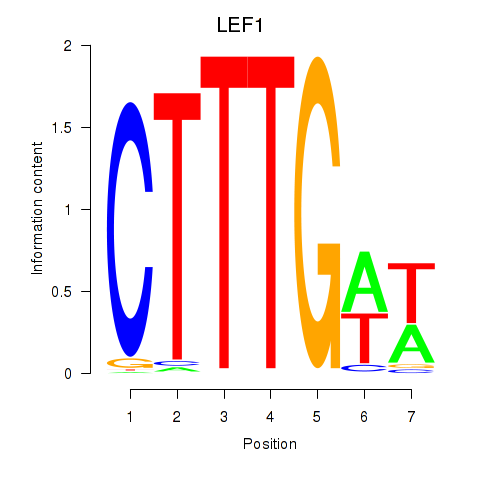
LEF1
|
ENSG00000138795.10 | LEF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg38_v1_chr4_-_108167784_108167890, hg38_v1_chr4_-_108166715_108166726 | -0.29 | 1.1e-01 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_138179093 | 1.72 |
ENST00000394894.8
|
KIF20A
|
kinesin family member 20A |
| chr5_+_138179145 | 1.72 |
ENST00000508792.5
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr3_+_19148500 | 1.67 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr2_-_1744442 | 1.66 |
ENST00000433670.5
ENST00000425171.1 ENST00000252804.9 |
PXDN
|
peroxidasin |
| chr3_-_142029108 | 1.60 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr3_-_71130557 | 1.42 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1 |
| chr14_+_54397021 | 1.35 |
ENST00000541304.5
|
CDKN3
|
cyclin dependent kinase inhibitor 3 |
| chr7_-_41703062 | 1.32 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr14_+_54396949 | 1.31 |
ENST00000611205.4
|
CDKN3
|
cyclin dependent kinase inhibitor 3 |
| chrX_+_38561530 | 1.31 |
ENST00000378482.7
ENST00000286824.6 |
TSPAN7
|
tetraspanin 7 |
| chr3_-_112641292 | 1.30 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr14_+_54396964 | 1.30 |
ENST00000543789.6
ENST00000442975.6 ENST00000458126.6 ENST00000556102.6 ENST00000335183.11 |
CDKN3
|
cyclin dependent kinase inhibitor 3 |
| chr3_-_71130963 | 1.25 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr1_-_205449924 | 1.23 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr9_+_98943898 | 1.19 |
ENST00000375001.8
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr7_+_120988683 | 1.09 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr3_-_33645253 | 1.09 |
ENST00000333778.10
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_112641128 | 1.08 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr1_-_205455954 | 1.07 |
ENST00000495594.2
ENST00000629624.2 |
LEMD1
BLACAT1
|
LEM domain containing 1 bladder cancer associated transcript 1 |
| chr9_+_98943705 | 1.01 |
ENST00000610452.1
|
COL15A1
|
collagen type XV alpha 1 chain |
| chr5_-_147453888 | 1.00 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr20_+_38926312 | 1.00 |
ENST00000619304.4
ENST00000619850.2 |
FAM83D
|
family with sequence similarity 83 member D |
| chr1_+_81800368 | 1.00 |
ENST00000674489.1
ENST00000674442.1 ENST00000674419.1 ENST00000674407.1 ENST00000674168.1 ENST00000674307.1 ENST00000674209.1 ENST00000370715.5 ENST00000370713.5 ENST00000319517.10 ENST00000627151.2 ENST00000370717.6 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr11_-_75306688 | 0.97 |
ENST00000532525.1
|
ARRB1
|
arrestin beta 1 |
| chr7_-_148884266 | 0.96 |
ENST00000483967.5
ENST00000320356.7 |
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chrX_-_48470243 | 0.92 |
ENST00000429543.2
ENST00000620913.5 |
SLC38A5
|
solute carrier family 38 member 5 |
| chr15_-_74202742 | 0.91 |
ENST00000395105.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chrX_-_48470163 | 0.91 |
ENST00000595796.5
|
SLC38A5
|
solute carrier family 38 member 5 |
| chr7_-_148884159 | 0.90 |
ENST00000478654.5
ENST00000460911.5 ENST00000350995.6 |
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr9_-_23826299 | 0.89 |
ENST00000380117.5
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr8_-_123541197 | 0.89 |
ENST00000517956.5
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr5_+_162068031 | 0.81 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chrY_-_13479938 | 0.80 |
ENST00000382893.2
ENST00000382896.9 ENST00000545955.6 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr1_-_226309071 | 0.80 |
ENST00000359525.2
ENST00000681046.1 ENST00000460719.5 |
LIN9
|
lin-9 DREAM MuvB core complex component |
| chr5_+_162067858 | 0.80 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067990 | 0.78 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr3_+_39467672 | 0.77 |
ENST00000436143.6
ENST00000441980.6 ENST00000682069.1 ENST00000311042.10 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr10_+_52314272 | 0.77 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_+_232870284 | 0.75 |
ENST00000331342.4
|
SNORC
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr6_+_99606833 | 0.75 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13 |
| chr7_+_28412511 | 0.75 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr3_-_71304741 | 0.74 |
ENST00000484350.5
|
FOXP1
|
forkhead box P1 |
| chr3_-_71130892 | 0.72 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr9_+_113876282 | 0.71 |
ENST00000374126.9
ENST00000615615.4 ENST00000288466.11 |
ZNF618
|
zinc finger protein 618 |
| chr5_+_162067500 | 0.71 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_55160161 | 0.69 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr21_+_29299368 | 0.67 |
ENST00000399921.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr2_-_25982471 | 0.67 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr6_-_63319943 | 0.66 |
ENST00000622415.1
ENST00000370658.9 ENST00000485906.6 ENST00000370657.9 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr9_-_91423819 | 0.64 |
ENST00000297689.4
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr4_+_1871373 | 0.63 |
ENST00000508803.6
ENST00000507820.5 ENST00000514045.5 |
NSD2
|
nuclear receptor binding SET domain protein 2 |
| chr14_+_85530127 | 0.62 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_+_55174508 | 0.62 |
ENST00000370862.4
|
HCRTR2
|
hypocretin receptor 2 |
| chr19_-_49361475 | 0.60 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2 |
| chr15_-_101252040 | 0.60 |
ENST00000254190.4
|
CHSY1
|
chondroitin sulfate synthase 1 |
| chr9_+_74615582 | 0.60 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr5_-_157575767 | 0.60 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr7_+_30852273 | 0.59 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough |
| chr15_-_55917080 | 0.59 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr10_+_112950757 | 0.59 |
ENST00000629706.2
ENST00000346198.5 |
TCF7L2
|
transcription factor 7 like 2 |
| chr7_+_130207847 | 0.59 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr5_-_65722094 | 0.59 |
ENST00000381007.9
|
SGTB
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr3_+_141386393 | 0.58 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_+_5592805 | 0.58 |
ENST00000382361.8
|
FSCN1
|
fascin actin-bundling protein 1 |
| chr1_+_84181630 | 0.58 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr7_-_25228485 | 0.58 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr12_-_16606795 | 0.56 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr3_+_39467598 | 0.56 |
ENST00000428261.5
ENST00000420739.5 ENST00000415443.5 ENST00000447324.5 ENST00000383754.7 |
MOBP
|
myelin associated oligodendrocyte basic protein |
| chr5_+_172641241 | 0.56 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr5_+_31193678 | 0.56 |
ENST00000265071.3
|
CDH6
|
cadherin 6 |
| chrX_+_96684712 | 0.56 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2 |
| chr12_+_32107296 | 0.55 |
ENST00000551086.1
|
BICD1
|
BICD cargo adaptor 1 |
| chr19_-_49362376 | 0.55 |
ENST00000601519.5
ENST00000593945.6 ENST00000539846.5 ENST00000596757.1 ENST00000311227.6 |
TEAD2
|
TEA domain transcription factor 2 |
| chr2_+_11539833 | 0.55 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr2_-_160200289 | 0.54 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr2_-_72147819 | 0.54 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr3_+_183265302 | 0.53 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr4_-_138242325 | 0.53 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr9_+_125748175 | 0.53 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr16_-_1884231 | 0.53 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chr16_-_46831043 | 0.52 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_-_71581540 | 0.51 |
ENST00000650068.1
|
FOXP1
|
forkhead box P1 |
| chrX_+_47078380 | 0.51 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr7_-_27180230 | 0.51 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr14_+_75280078 | 0.50 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr17_-_41481140 | 0.50 |
ENST00000246639.6
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chrX_+_47078330 | 0.50 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chr5_+_31193739 | 0.50 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr17_-_44947637 | 0.50 |
ENST00000587309.5
ENST00000593135.6 |
KIF18B
|
kinesin family member 18B |
| chr7_-_100100716 | 0.50 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr12_+_53097656 | 0.50 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr21_+_38256698 | 0.49 |
ENST00000613499.4
ENST00000612702.4 ENST00000398925.5 ENST00000398928.5 ENST00000328656.8 ENST00000443341.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr10_+_84452208 | 0.49 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr2_-_160200251 | 0.49 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chrX_+_47078434 | 0.49 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr5_-_161546671 | 0.48 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr5_-_138338325 | 0.48 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr10_-_99620401 | 0.48 |
ENST00000370495.6
|
SLC25A28
|
solute carrier family 25 member 28 |
| chr5_+_162067764 | 0.48 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_-_161546970 | 0.48 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr3_-_33645433 | 0.48 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_-_88175432 | 0.48 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.11 |
RAB38
|
RAB38, member RAS oncogene family |
| chrX_-_100928903 | 0.47 |
ENST00000372956.3
|
XKRX
|
XK related X-linked |
| chr10_+_112950586 | 0.47 |
ENST00000355717.9
ENST00000352065.10 ENST00000369395.6 ENST00000545257.6 |
TCF7L2
|
transcription factor 7 like 2 |
| chr2_-_160200310 | 0.47 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr6_-_152983031 | 0.47 |
ENST00000229758.8
|
FBXO5
|
F-box protein 5 |
| chr18_+_44697118 | 0.47 |
ENST00000677077.1
|
SETBP1
|
SET binding protein 1 |
| chr5_+_162067458 | 0.47 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr3_-_142028597 | 0.46 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chr6_-_25874212 | 0.46 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr13_+_29428603 | 0.46 |
ENST00000380808.6
|
MTUS2
|
microtubule associated scaffold protein 2 |
| chr2_-_215393126 | 0.46 |
ENST00000456923.5
|
FN1
|
fibronectin 1 |
| chr10_-_67838173 | 0.45 |
ENST00000225171.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr7_+_74289397 | 0.45 |
ENST00000223398.11
ENST00000361545.9 |
CLIP2
|
CAP-Gly domain containing linker protein 2 |
| chr14_+_60734727 | 0.45 |
ENST00000261245.9
ENST00000539616.6 |
MNAT1
|
MNAT1 component of CDK activating kinase |
| chr21_-_30813270 | 0.44 |
ENST00000329621.6
|
KRTAP8-1
|
keratin associated protein 8-1 |
| chr7_-_19773569 | 0.44 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr11_-_41459592 | 0.44 |
ENST00000528697.6
ENST00000530763.5 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr10_-_114404480 | 0.43 |
ENST00000419268.1
ENST00000304129.9 |
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr3_-_71064915 | 0.43 |
ENST00000614176.5
ENST00000485326.7 |
FOXP1
|
forkhead box P1 |
| chr10_-_114404756 | 0.43 |
ENST00000369271.7
|
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr2_+_96325294 | 0.43 |
ENST00000439118.3
ENST00000420176.5 ENST00000536814.1 |
ITPRIPL1
|
ITPRIP like 1 |
| chr11_-_57530714 | 0.43 |
ENST00000525158.1
ENST00000257245.9 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 |
| chr17_+_82458174 | 0.43 |
ENST00000579198.5
ENST00000390006.8 ENST00000580296.5 |
NARF
|
nuclear prelamin A recognition factor |
| chr13_-_23433676 | 0.43 |
ENST00000682547.1
ENST00000455470.6 ENST00000382292.9 |
SACS
|
sacsin molecular chaperone |
| chr1_-_53838276 | 0.43 |
ENST00000371429.4
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr3_-_71064964 | 0.43 |
ENST00000650387.1
|
FOXP1
|
forkhead box P1 |
| chr8_+_62248933 | 0.42 |
ENST00000523211.5
ENST00000524201.1 |
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr18_-_48950960 | 0.42 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chrX_+_96684801 | 0.42 |
ENST00000324765.13
|
DIAPH2
|
diaphanous related formin 2 |
| chr5_+_3595820 | 0.42 |
ENST00000302006.4
|
IRX1
|
iroquois homeobox 1 |
| chr12_+_1629197 | 0.42 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr21_+_38256984 | 0.41 |
ENST00000398938.7
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr4_+_94207596 | 0.41 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_+_82458649 | 0.41 |
ENST00000345415.11
ENST00000412079.6 ENST00000457415.7 ENST00000584411.5 ENST00000577432.5 |
NARF
|
nuclear prelamin A recognition factor |
| chr14_-_34713759 | 0.41 |
ENST00000673315.1
|
CFL2
|
cofilin 2 |
| chr16_-_9943182 | 0.40 |
ENST00000535259.6
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr6_-_152983561 | 0.40 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr15_-_70096604 | 0.40 |
ENST00000559048.5
ENST00000560939.5 ENST00000440567.7 ENST00000557907.5 ENST00000558379.5 ENST00000559929.5 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr7_-_29195186 | 0.40 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr2_+_27442365 | 0.40 |
ENST00000543753.5
ENST00000288873.7 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr15_-_55917129 | 0.39 |
ENST00000338963.6
ENST00000508342.5 |
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr11_+_112961480 | 0.39 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr15_-_70096260 | 0.39 |
ENST00000558201.5
|
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr1_+_84164684 | 0.39 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_-_16608183 | 0.39 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr11_-_66677748 | 0.39 |
ENST00000525754.5
ENST00000531969.5 ENST00000524637.1 ENST00000531036.2 ENST00000310046.9 |
RBM4B
|
RNA binding motif protein 4B |
| chr11_+_112961402 | 0.39 |
ENST00000613217.4
ENST00000316851.12 ENST00000620046.4 ENST00000531044.5 ENST00000529356.5 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr10_-_17129786 | 0.39 |
ENST00000377833.10
|
CUBN
|
cubilin |
| chr14_-_23578756 | 0.38 |
ENST00000397118.7
ENST00000356300.9 |
JPH4
|
junctophilin 4 |
| chr1_+_93079234 | 0.38 |
ENST00000540243.5
ENST00000545708.5 |
MTF2
|
metal response element binding transcription factor 2 |
| chr12_-_16608073 | 0.38 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr7_+_73830988 | 0.38 |
ENST00000340958.4
|
CLDN4
|
claudin 4 |
| chr1_-_153113507 | 0.38 |
ENST00000468739.2
|
SPRR2F
|
small proline rich protein 2F |
| chr14_+_85530163 | 0.38 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chrX_+_54809060 | 0.37 |
ENST00000396224.1
|
MAGED2
|
MAGE family member D2 |
| chr12_+_54561442 | 0.37 |
ENST00000550620.1
|
PDE1B
|
phosphodiesterase 1B |
| chr2_+_170715317 | 0.37 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chr5_+_171419635 | 0.37 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr2_+_27442421 | 0.37 |
ENST00000407293.5
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr13_-_23433735 | 0.37 |
ENST00000423156.2
ENST00000683210.1 ENST00000682775.1 ENST00000684497.1 ENST00000682944.1 ENST00000683489.1 ENST00000684385.1 ENST00000683680.1 |
SACS
|
sacsin molecular chaperone |
| chr6_+_113857333 | 0.36 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr16_-_22374580 | 0.36 |
ENST00000268383.7
|
CDR2
|
cerebellar degeneration related protein 2 |
| chr1_-_156572535 | 0.36 |
ENST00000361170.7
|
IQGAP3
|
IQ motif containing GTPase activating protein 3 |
| chr14_-_80959484 | 0.36 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chrX_-_43973382 | 0.35 |
ENST00000642620.1
ENST00000647044.1 |
NDP
|
norrin cystine knot growth factor NDP |
| chr19_+_751104 | 0.35 |
ENST00000215582.8
|
MISP
|
mitotic spindle positioning |
| chr9_+_100427254 | 0.35 |
ENST00000374885.5
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr4_+_153466324 | 0.34 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr5_+_161848536 | 0.34 |
ENST00000519621.2
ENST00000636573.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr18_-_26865732 | 0.34 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr11_+_5389377 | 0.34 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chrX_+_96684638 | 0.34 |
ENST00000355827.8
ENST00000373061.7 |
DIAPH2
|
diaphanous related formin 2 |
| chr4_+_94207845 | 0.34 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_-_41055211 | 0.34 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr2_+_203867943 | 0.33 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chrX_+_96684929 | 0.33 |
ENST00000373054.5
|
DIAPH2
|
diaphanous related formin 2 |
| chr12_-_31324129 | 0.33 |
ENST00000454658.6
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr1_+_47333863 | 0.33 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine/uridine monophosphate kinase 1 |
| chr6_+_20403679 | 0.33 |
ENST00000535432.2
|
E2F3
|
E2F transcription factor 3 |
| chr8_-_17002327 | 0.33 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20 |
| chr12_+_103965835 | 0.33 |
ENST00000266775.13
ENST00000544861.5 |
TDG
|
thymine DNA glycosylase |
| chr12_-_117968972 | 0.32 |
ENST00000339824.7
|
KSR2
|
kinase suppressor of ras 2 |
| chrX_-_81201886 | 0.32 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr8_-_132760548 | 0.32 |
ENST00000519187.5
ENST00000523829.5 ENST00000677595.1 ENST00000356838.7 ENST00000377901.8 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr2_-_165203870 | 0.32 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr3_-_147406520 | 0.32 |
ENST00000463250.1
ENST00000383075.8 |
ZIC4
|
Zic family member 4 |
| chr3_+_107525378 | 0.32 |
ENST00000456419.5
ENST00000402163.6 |
BBX
|
BBX high mobility group box domain containing |
| chr2_-_169362527 | 0.32 |
ENST00000649046.1
ENST00000443831.1 |
LRP2
|
LDL receptor related protein 2 |
| chr17_+_77451244 | 0.32 |
ENST00000591088.5
|
SEPTIN9
|
septin 9 |
| chr12_+_103965798 | 0.32 |
ENST00000436021.6
|
TDG
|
thymine DNA glycosylase |
| chr13_-_27969295 | 0.32 |
ENST00000381020.8
|
CDX2
|
caudal type homeobox 2 |
| chr11_-_57427474 | 0.32 |
ENST00000533235.5
ENST00000526621.5 ENST00000352187.5 |
SLC43A3
|
solute carrier family 43 member 3 |
| chr18_-_26865689 | 0.32 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr7_+_69599588 | 0.31 |
ENST00000403018.3
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr3_-_149086488 | 0.31 |
ENST00000392912.6
ENST00000465259.5 ENST00000310053.10 ENST00000494055.5 |
HLTF
|
helicase like transcription factor |
| chr6_+_37170133 | 0.31 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr7_+_80646305 | 0.31 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr11_+_112961247 | 0.31 |
ENST00000621518.4
ENST00000618266.4 ENST00000615112.4 ENST00000615285.4 ENST00000619839.4 ENST00000401611.6 ENST00000621128.4 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr13_+_75636311 | 0.31 |
ENST00000377499.9
ENST00000377534.8 |
LMO7
|
LIM domain 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.4 | 1.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 1.5 | GO:1903611 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.3 | 1.0 | GO:0019089 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.3 | 5.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 0.9 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.3 | 0.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 0.8 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.9 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.2 | 0.7 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 5.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.6 | GO:1990009 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.2 | 1.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 | 2.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.5 | GO:0097086 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.2 | 2.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 0.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.5 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.6 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.2 | 2.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 1.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 0.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 3.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.4 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.7 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.5 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.9 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.6 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.4 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0032600 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.3 | GO:0019418 | succinate transport(GO:0015744) sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) succinate transmembrane transport(GO:0071422) |
| 0.1 | 0.1 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 0.9 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.1 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.6 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.2 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 2.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.5 | GO:0097491 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.5 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 1.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.8 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.5 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.4 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 4.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:2000174 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.3 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.0 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0048619 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 2.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.9 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 1.0 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.0 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 1.4 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 2.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.0 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 2.5 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.2 | 2.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.6 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 5.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.4 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 2.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0005816 | spindle pole body(GO:0005816) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.4 | 1.5 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.3 | 1.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 0.9 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 1.0 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 4.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.5 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 1.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.8 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.8 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 4.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 5.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 4.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.0 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |