Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for LHX3
Z-value: 0.46
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.17 | LHX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | hg38_v1_chr9_-_136203183_136203194, hg38_v1_chr9_-_136205122_136205133 | -0.19 | 3.1e-01 | Click! |
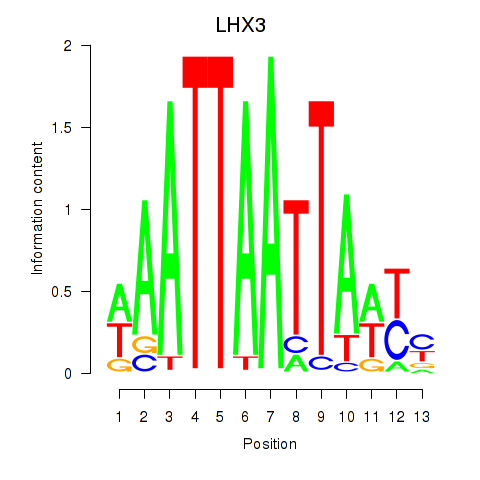
Activity profile of LHX3 motif
Sorted Z-values of LHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_96928310 | 0.91 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr12_-_10130241 | 0.70 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr12_-_10130143 | 0.66 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr12_-_10130082 | 0.60 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr17_-_40937445 | 0.55 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr17_-_40937641 | 0.54 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr19_+_11346556 | 0.53 |
ENST00000587531.5
|
CCDC159
|
coiled-coil domain containing 159 |
| chr9_-_72953047 | 0.52 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr3_+_41194741 | 0.50 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr1_-_111488795 | 0.49 |
ENST00000472933.2
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr5_+_148312416 | 0.45 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr9_+_122370523 | 0.43 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr11_-_63608542 | 0.43 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr8_-_109680812 | 0.42 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr19_+_48695952 | 0.38 |
ENST00000522966.2
ENST00000425340.3 ENST00000391876.5 |
FUT2
|
fucosyltransferase 2 |
| chr11_+_26994102 | 0.32 |
ENST00000318627.4
|
FIBIN
|
fin bud initiation factor homolog |
| chr19_+_49513154 | 0.30 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chrX_-_13817027 | 0.28 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr16_-_28623560 | 0.28 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr19_+_49513353 | 0.28 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr9_+_72577369 | 0.27 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr1_+_81306096 | 0.27 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr19_-_48646155 | 0.27 |
ENST00000084798.9
|
CA11
|
carbonic anhydrase 11 |
| chr18_-_55423757 | 0.26 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr9_+_72577788 | 0.26 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr15_-_55408245 | 0.26 |
ENST00000563171.5
ENST00000425574.7 ENST00000442196.8 ENST00000564092.1 |
CCPG1
|
cell cycle progression 1 |
| chr10_+_87357720 | 0.25 |
ENST00000412718.3
ENST00000381697.7 |
NUTM2D
|
NUT family member 2D |
| chr8_-_109974688 | 0.25 |
ENST00000297404.1
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr5_+_68290637 | 0.25 |
ENST00000336483.9
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_-_158426237 | 0.24 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr4_-_23890035 | 0.24 |
ENST00000507380.1
ENST00000264867.7 |
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr1_+_244051275 | 0.24 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr16_-_28623330 | 0.24 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr17_+_1771688 | 0.24 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr19_+_14583076 | 0.24 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr18_-_55635948 | 0.23 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr1_+_115029823 | 0.22 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chrX_+_109535775 | 0.22 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr2_+_26401909 | 0.22 |
ENST00000288710.7
|
DRC1
|
dynein regulatory complex subunit 1 |
| chr9_+_122371014 | 0.21 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr4_+_186069144 | 0.21 |
ENST00000513189.1
ENST00000296795.8 |
TLR3
|
toll like receptor 3 |
| chr9_+_79573162 | 0.21 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr15_+_64387828 | 0.20 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr18_+_74534594 | 0.20 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 |
| chr1_-_182671902 | 0.20 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8 |
| chr18_+_74534493 | 0.20 |
ENST00000358821.8
|
CNDP1
|
carnosine dipeptidase 1 |
| chr2_+_233917371 | 0.19 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr1_-_182671853 | 0.19 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr15_-_75455767 | 0.19 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_96505345 | 0.19 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr20_+_6007245 | 0.19 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr9_-_113303271 | 0.18 |
ENST00000297894.5
ENST00000489339.2 |
RNF183
|
ring finger protein 183 |
| chr1_-_56819365 | 0.17 |
ENST00000343433.7
|
FYB2
|
FYN binding protein 2 |
| chr1_-_48400826 | 0.17 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr8_+_106726115 | 0.17 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr7_+_90383672 | 0.17 |
ENST00000416322.5
|
CLDN12
|
claudin 12 |
| chr13_-_49401497 | 0.17 |
ENST00000457041.5
ENST00000355854.8 |
CAB39L
|
calcium binding protein 39 like |
| chr10_-_27240505 | 0.17 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr12_-_118190510 | 0.17 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_141177790 | 0.17 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr19_-_43465596 | 0.17 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr7_-_36724543 | 0.16 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr7_-_36724457 | 0.16 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr10_+_47918659 | 0.16 |
ENST00000576178.6
ENST00000613306.1 |
NPY4R2
|
neuropeptide Y receptor Y4-2 |
| chr19_-_11346486 | 0.16 |
ENST00000590482.5
|
TMEM205
|
transmembrane protein 205 |
| chr3_-_191282383 | 0.15 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr12_-_52367478 | 0.15 |
ENST00000257901.7
|
KRT85
|
keratin 85 |
| chr2_-_98663464 | 0.15 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chrM_-_14669 | 0.14 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr9_+_69145463 | 0.14 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2 |
| chr3_-_132684685 | 0.14 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr17_-_10469558 | 0.14 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chrX_+_106168297 | 0.13 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chr6_-_49744378 | 0.13 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr20_+_31475278 | 0.13 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1 |
| chr6_-_49744434 | 0.12 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr8_-_18083278 | 0.12 |
ENST00000636691.1
|
ASAH1
|
N-acylsphingosine amidohydrolase 1 |
| chr9_+_72577939 | 0.12 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr11_-_129024157 | 0.12 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_+_104339796 | 0.11 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr14_+_61697622 | 0.11 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr20_+_45416551 | 0.11 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr20_-_31475125 | 0.11 |
ENST00000317676.3
|
DEFB124
|
defensin beta 124 |
| chr17_+_70075317 | 0.11 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr19_+_11346499 | 0.11 |
ENST00000458408.6
ENST00000586451.5 ENST00000588592.5 |
CCDC159
|
coiled-coil domain containing 159 |
| chr8_-_79767462 | 0.10 |
ENST00000674295.1
ENST00000518733.1 ENST00000674418.1 ENST00000674358.1 ENST00000354724.8 |
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1 |
| chr10_-_114144599 | 0.10 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr8_-_131040890 | 0.10 |
ENST00000286355.10
|
ADCY8
|
adenylate cyclase 8 |
| chr2_-_99255107 | 0.10 |
ENST00000333017.6
ENST00000626374.2 ENST00000409679.5 ENST00000423306.1 |
LYG2
|
lysozyme g2 |
| chr7_+_92057602 | 0.10 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr3_-_142000353 | 0.10 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr3_+_46877705 | 0.10 |
ENST00000449590.6
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr4_-_86357722 | 0.09 |
ENST00000641341.1
ENST00000642038.1 ENST00000641116.1 ENST00000641767.1 ENST00000639242.1 ENST00000638313.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_+_67570741 | 0.09 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr17_-_66229380 | 0.08 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr7_+_107583919 | 0.08 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr1_-_23799561 | 0.08 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr4_+_53377630 | 0.07 |
ENST00000337488.11
ENST00000358575.9 ENST00000507922.5 ENST00000306932.10 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr10_-_88851809 | 0.07 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr15_+_48191648 | 0.07 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr6_+_72216442 | 0.07 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr18_-_3845321 | 0.07 |
ENST00000539435.5
ENST00000400147.6 |
DLGAP1
|
DLG associated protein 1 |
| chr10_+_18260715 | 0.07 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr12_-_86256299 | 0.07 |
ENST00000552808.6
ENST00000547225.5 |
MGAT4C
|
MGAT4 family member C |
| chr8_-_25424260 | 0.06 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1 |
| chr22_+_23145366 | 0.06 |
ENST00000341989.9
ENST00000263116.8 |
RAB36
|
RAB36, member RAS oncogene family |
| chr17_+_46511511 | 0.06 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chrX_+_85003863 | 0.06 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr12_+_49346911 | 0.06 |
ENST00000395069.3
|
DNAJC22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr14_+_19743571 | 0.06 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr5_+_55853314 | 0.06 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chr3_-_47475811 | 0.06 |
ENST00000265565.10
ENST00000428413.5 |
SCAP
|
SREBF chaperone |
| chr18_+_58341038 | 0.05 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr19_-_6279921 | 0.05 |
ENST00000252674.9
|
MLLT1
|
MLLT1 super elongation complex subunit |
| chr7_+_138460238 | 0.05 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr6_-_159045104 | 0.05 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr4_+_112647059 | 0.05 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr4_+_41935114 | 0.05 |
ENST00000508448.5
ENST00000513702.5 ENST00000325094.9 |
TMEM33
|
transmembrane protein 33 |
| chr3_-_12545499 | 0.04 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr3_+_69763726 | 0.04 |
ENST00000448226.9
|
MITF
|
melanocyte inducing transcription factor |
| chr5_+_141150012 | 0.04 |
ENST00000231136.4
ENST00000622991.1 |
PCDHB6
|
protocadherin beta 6 |
| chr8_-_56211257 | 0.04 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr3_+_158110052 | 0.04 |
ENST00000295930.7
ENST00000471994.5 ENST00000482822.3 ENST00000476899.6 ENST00000683899.1 ENST00000684604.1 ENST00000682164.1 ENST00000464171.5 ENST00000611884.5 ENST00000312179.10 ENST00000475278.6 |
RSRC1
|
arginine and serine rich coiled-coil 1 |
| chr18_-_24272179 | 0.04 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr1_-_23799533 | 0.03 |
ENST00000429356.5
|
GALE
|
UDP-galactose-4-epimerase |
| chr2_+_134838610 | 0.03 |
ENST00000356140.10
ENST00000392928.5 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_+_70592253 | 0.03 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr4_+_41613476 | 0.03 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chrM_+_10055 | 0.03 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr16_+_28553908 | 0.03 |
ENST00000317058.8
|
SGF29
|
SAGA complex associated factor 29 |
| chr9_+_122371036 | 0.03 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr7_-_25228485 | 0.03 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr2_-_187448244 | 0.02 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr5_-_79514127 | 0.02 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr16_+_15395745 | 0.02 |
ENST00000287594.7
ENST00000396385.4 ENST00000568766.1 |
MPV17L
ENSG00000261130.5
|
MPV17 mitochondrial inner membrane protein like novel protein |
| chr11_-_790062 | 0.02 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_-_92486916 | 0.02 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr19_+_9185594 | 0.02 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr6_+_82363199 | 0.02 |
ENST00000535040.4
|
TPBG
|
trophoblast glycoprotein |
| chr7_-_123199960 | 0.01 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr3_-_20012250 | 0.01 |
ENST00000389050.5
|
PP2D1
|
protein phosphatase 2C like domain containing 1 |
| chr5_-_16916400 | 0.01 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr6_+_130018565 | 0.01 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr3_-_165078480 | 0.01 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr7_+_30145789 | 0.01 |
ENST00000324489.5
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr1_-_230869564 | 0.01 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr17_-_44066595 | 0.01 |
ENST00000585388.2
ENST00000293406.8 |
LSM12
|
LSM12 homolog |
| chr8_+_91249307 | 0.00 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr4_+_157220691 | 0.00 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr1_+_15341744 | 0.00 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.5 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 2.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:2000182 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.4 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0036304 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 2.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |