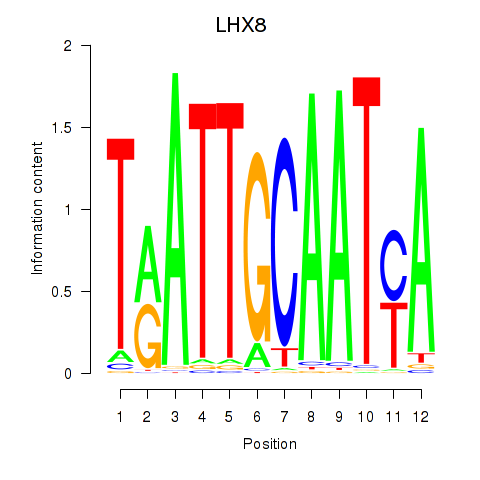
|
chr3_-_112975018
Show fit
|
1.33 |
ENST00000471858.5
ENST00000308611.8
ENST00000295863.4
|
CD200R1
|
CD200 receptor 1
|
|
chr16_-_53703883
Show fit
|
1.13 |
ENST00000262135.9
ENST00000564374.5
ENST00000566096.5
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr16_-_53703810
Show fit
|
1.10 |
ENST00000569716.1
ENST00000562588.5
ENST00000621565.5
ENST00000562230.5
ENST00000563746.5
ENST00000568653.7
ENST00000647211.2
|
RPGRIP1L
|
RPGRIP1 like
|
|
chr11_+_102047422
Show fit
|
1.08 |
ENST00000434758.7
ENST00000526781.5
ENST00000534360.1
|
CFAP300
|
cilia and flagella associated protein 300
|
|
chr3_-_112974912
Show fit
|
0.95 |
ENST00000440122.6
ENST00000490004.1
|
CD200R1
|
CD200 receptor 1
|
|
chr22_-_50532489
Show fit
|
0.90 |
ENST00000329363.9
ENST00000437588.2
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr8_+_17497108
Show fit
|
0.84 |
ENST00000470360.5
|
SLC7A2
|
solute carrier family 7 member 2
|
|
chr2_-_224402097
Show fit
|
0.83 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B
|
|
chr11_-_5227063
Show fit
|
0.82 |
ENST00000335295.4
ENST00000485743.1
ENST00000647020.1
|
HBB
|
hemoglobin subunit beta
|
|
chr3_-_112829367
Show fit
|
0.78 |
ENST00000448932.4
ENST00000617549.3
|
CD200R1L
|
CD200 receptor 1 like
|
|
chr11_-_114595777
Show fit
|
0.74 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4
|
|
chr8_+_17497078
Show fit
|
0.71 |
ENST00000494857.6
ENST00000522656.5
|
SLC7A2
|
solute carrier family 7 member 2
|
|
chr14_-_91946989
Show fit
|
0.66 |
ENST00000556154.5
|
FBLN5
|
fibulin 5
|
|
chr12_-_71157992
Show fit
|
0.62 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8
|
|
chr12_-_71157872
Show fit
|
0.60 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8
|
|
chr12_-_7891140
Show fit
|
0.59 |
ENST00000539924.5
|
SLC2A14
|
solute carrier family 2 member 14
|
|
chr7_+_90245207
Show fit
|
0.57 |
ENST00000497910.5
|
CFAP69
|
cilia and flagella associated protein 69
|
|
chr4_+_69995958
Show fit
|
0.51 |
ENST00000381060.2
ENST00000246895.9
|
STATH
|
statherin
|
|
chr6_+_29396555
Show fit
|
0.50 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2
|
|
chr1_-_48472166
Show fit
|
0.50 |
ENST00000371847.8
ENST00000396199.7
|
SPATA6
|
spermatogenesis associated 6
|
|
chr8_+_98064522
Show fit
|
0.45 |
ENST00000545282.1
|
ERICH5
|
glutamate rich 5
|
|
chr16_+_22206255
Show fit
|
0.44 |
ENST00000263026.10
|
EEF2K
|
eukaryotic elongation factor 2 kinase
|
|
chr12_-_39340963
Show fit
|
0.43 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A
|
|
chr2_-_151261839
Show fit
|
0.43 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43
|
|
chr5_-_75712419
Show fit
|
0.38 |
ENST00000510798.5
ENST00000446329.6
|
POC5
|
POC5 centriolar protein
|
|
chr12_-_54588516
Show fit
|
0.38 |
ENST00000547431.5
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A
|
|
chr6_+_29301701
Show fit
|
0.38 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1
|
|
chr5_+_140786136
Show fit
|
0.38 |
ENST00000378133.4
ENST00000504120.4
|
PCDHA1
|
protocadherin alpha 1
|
|
chr1_-_217089627
Show fit
|
0.38 |
ENST00000361525.7
|
ESRRG
|
estrogen related receptor gamma
|
|
chr7_+_114414809
Show fit
|
0.38 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2
|
|
chr19_-_10194898
Show fit
|
0.37 |
ENST00000359526.9
ENST00000679103.1
ENST00000676610.1
ENST00000678804.1
ENST00000679313.1
ENST00000677946.1
|
DNMT1
|
DNA methyltransferase 1
|
|
chr4_-_79326008
Show fit
|
0.37 |
ENST00000286794.5
|
NAA11
|
N-alpha-acetyltransferase 11, NatA catalytic subunit
|
|
chr6_+_29306626
Show fit
|
0.36 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1
|
|
chr8_+_98064559
Show fit
|
0.34 |
ENST00000318528.8
|
ERICH5
|
glutamate rich 5
|
|
chr12_+_63779894
Show fit
|
0.34 |
ENST00000261234.11
|
RXYLT1
|
ribitol xylosyltransferase 1
|
|
chr1_-_236281951
Show fit
|
0.33 |
ENST00000354619.10
|
ERO1B
|
endoplasmic reticulum oxidoreductase 1 beta
|
|
chr2_-_151973780
Show fit
|
0.33 |
ENST00000637514.1
ENST00000636350.1
ENST00000434468.2
ENST00000637762.1
ENST00000637779.1
ENST00000637547.1
ENST00000636901.1
ENST00000397327.7
ENST00000636721.1
ENST00000636380.1
ENST00000637284.1
ENST00000636617.1
ENST00000636947.1
ENST00000638091.1
ENST00000636108.1
ENST00000638040.1
ENST00000636773.1
ENST00000637418.1
ENST00000637216.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4
|
|
chr6_-_49713564
Show fit
|
0.32 |
ENST00000616725.4
ENST00000618917.4
|
CRISP2
|
cysteine rich secretory protein 2
|
|
chr1_+_240014319
Show fit
|
0.32 |
ENST00000447095.5
|
FMN2
|
formin 2
|
|
chr1_+_212035717
Show fit
|
0.32 |
ENST00000366991.5
|
DTL
|
denticleless E3 ubiquitin protein ligase homolog
|
|
chr9_-_13165442
Show fit
|
0.32 |
ENST00000542239.1
ENST00000538841.5
ENST00000433359.6
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component
|
|
chr16_-_88703611
Show fit
|
0.31 |
ENST00000541206.6
|
RNF166
|
ring finger protein 166
|
|
chr19_+_21505656
Show fit
|
0.29 |
ENST00000618549.1
|
ZNF429
|
zinc finger protein 429
|
|
chr2_-_201739205
Show fit
|
0.29 |
ENST00000681558.1
ENST00000681495.1
|
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2
|
|
chr3_+_100609594
Show fit
|
0.28 |
ENST00000273352.8
|
ADGRG7
|
adhesion G protein-coupled receptor G7
|
|
chr6_-_149890900
Show fit
|
0.27 |
ENST00000529948.1
ENST00000357183.8
ENST00000367363.3
|
RAET1E
|
retinoic acid early transcript 1E
|
|
chr6_+_116369837
Show fit
|
0.27 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase
|
|
chr4_+_70242583
Show fit
|
0.27 |
ENST00000304954.3
|
CSN3
|
casein kappa
|
|
chr15_-_39782794
Show fit
|
0.25 |
ENST00000350221.4
|
FSIP1
|
fibrous sheath interacting protein 1
|
|
chr7_+_114414997
Show fit
|
0.25 |
ENST00000462331.5
ENST00000393491.7
ENST00000403559.8
ENST00000408937.7
ENST00000393498.6
ENST00000393495.7
ENST00000378237.7
|
FOXP2
|
forkhead box P2
|
|
chr4_-_109801978
Show fit
|
0.24 |
ENST00000510800.1
ENST00000512148.5
ENST00000394634.7
ENST00000394635.8
ENST00000645635.1
|
CFI
ENSG00000285330.1
|
complement factor I
novel protein
|
|
chr4_+_188139438
Show fit
|
0.23 |
ENST00000332517.4
|
TRIML1
|
tripartite motif family like 1
|
|
chr19_-_45424364
Show fit
|
0.23 |
ENST00000589165.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit
|
|
chr2_+_216081866
Show fit
|
0.23 |
ENST00000433112.5
ENST00000454545.5
ENST00000295658.8
ENST00000455479.5
ENST00000406027.2
ENST00000437356.7
|
TMEM169
|
transmembrane protein 169
|
|
chr10_+_133394094
Show fit
|
0.23 |
ENST00000477902.6
|
MTG1
|
mitochondrial ribosome associated GTPase 1
|
|
chr1_-_160523204
Show fit
|
0.23 |
ENST00000368055.1
ENST00000368057.8
ENST00000368059.7
|
SLAMF6
|
SLAM family member 6
|
|
chr8_+_74350394
Show fit
|
0.23 |
ENST00000675928.1
ENST00000676443.1
ENST00000676112.1
ENST00000674710.1
ENST00000434412.3
ENST00000675165.1
ENST00000674973.1
ENST00000220822.12
ENST00000675999.1
ENST00000523640.2
ENST00000524195.2
ENST00000675463.1
ENST00000676207.1
ENST00000674865.1
ENST00000676415.1
|
GDAP1
|
ganglioside induced differentiation associated protein 1
|
|
chr6_+_29395631
Show fit
|
0.22 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2
|
|
chr19_+_21505585
Show fit
|
0.22 |
ENST00000358491.9
ENST00000597078.5
|
ZNF429
|
zinc finger protein 429
|
|
chr12_+_53954870
Show fit
|
0.21 |
ENST00000243103.4
|
HOXC12
|
homeobox C12
|
|
chr19_+_47349298
Show fit
|
0.20 |
ENST00000328771.9
|
DHX34
|
DExH-box helicase 34
|
|
chr4_-_145180496
Show fit
|
0.20 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4
|
|
chr7_+_151341764
Show fit
|
0.20 |
ENST00000413040.7
ENST00000470229.6
ENST00000568733.6
|
NUB1
|
negative regulator of ubiquitin like proteins 1
|
|
chr10_+_92593112
Show fit
|
0.19 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11
|
|
chr11_-_84923403
Show fit
|
0.19 |
ENST00000532653.5
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr1_+_182450132
Show fit
|
0.19 |
ENST00000294854.13
|
RGSL1
|
regulator of G protein signaling like 1
|
|
chr6_+_154039480
Show fit
|
0.19 |
ENST00000229768.9
ENST00000419506.6
ENST00000524163.5
ENST00000414028.6
ENST00000435918.6
ENST00000337049.8
|
OPRM1
|
opioid receptor mu 1
|
|
chr11_-_84923162
Show fit
|
0.18 |
ENST00000524982.5
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr13_+_24309565
Show fit
|
0.17 |
ENST00000332018.4
|
C1QTNF9
|
C1q and TNF related 9
|
|
chr19_+_827823
Show fit
|
0.17 |
ENST00000233997.4
|
AZU1
|
azurocidin 1
|
|
chr3_+_195720974
Show fit
|
0.17 |
ENST00000447234.7
ENST00000320736.10
|
MUC20
|
mucin 20, cell surface associated
|
|
chr3_+_119468952
Show fit
|
0.17 |
ENST00000476573.5
ENST00000295588.9
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr4_-_94342826
Show fit
|
0.17 |
ENST00000295256.10
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr5_-_24644968
Show fit
|
0.17 |
ENST00000264463.8
|
CDH10
|
cadherin 10
|
|
chr9_+_26956384
Show fit
|
0.16 |
ENST00000518614.5
ENST00000380062.10
|
IFT74
|
intraflagellar transport 74
|
|
chr4_-_86101922
Show fit
|
0.16 |
ENST00000472236.5
ENST00000641881.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr7_-_7535863
Show fit
|
0.16 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain
|
|
chr19_-_39738017
Show fit
|
0.15 |
ENST00000221804.5
|
CLC
|
Charcot-Leyden crystal galectin
|
|
chr7_-_120858066
Show fit
|
0.15 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12
|
|
chr7_-_14841267
Show fit
|
0.15 |
ENST00000406247.7
ENST00000399322.7
|
DGKB
|
diacylglycerol kinase beta
|
|
chr19_+_44212525
Show fit
|
0.15 |
ENST00000391961.6
ENST00000621083.4
ENST00000313040.12
ENST00000586228.5
ENST00000588219.5
ENST00000589707.5
ENST00000588394.5
ENST00000589005.5
|
ZNF227
|
zinc finger protein 227
|
|
chr3_-_191282383
Show fit
|
0.15 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|
|
chr1_-_197067234
Show fit
|
0.15 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain
|
|
chr9_-_21228222
Show fit
|
0.14 |
ENST00000413767.2
|
IFNA17
|
interferon alpha 17
|
|
chr20_-_31870629
Show fit
|
0.14 |
ENST00000375966.8
ENST00000278979.7
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr20_+_31870628
Show fit
|
0.14 |
ENST00000375938.8
|
TTLL9
|
tubulin tyrosine ligase like 9
|
|
chr6_-_52763473
Show fit
|
0.13 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr9_-_15472732
Show fit
|
0.13 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr18_+_32091849
Show fit
|
0.13 |
ENST00000261593.8
ENST00000578914.1
|
RNF138
|
ring finger protein 138
|
|
chr7_+_64045420
Show fit
|
0.13 |
ENST00000647787.1
ENST00000456806.3
|
ENSG00000285544.1
ZNF727
|
novel transcript
zinc finger protein 727
|
|
chr3_+_42936354
Show fit
|
0.13 |
ENST00000383748.9
|
KRBOX1
|
KRAB box domain containing 1
|
|
chr2_-_177263522
Show fit
|
0.12 |
ENST00000448782.5
ENST00000446151.6
|
NFE2L2
|
nuclear factor, erythroid 2 like 2
|
|
chr15_+_65550819
Show fit
|
0.12 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3
|
|
chr9_-_179008
Show fit
|
0.12 |
ENST00000613508.4
ENST00000382447.8
ENST00000382389.5
ENST00000377447.7
ENST00000382393.2
ENST00000314367.14
ENST00000356521.8
ENST00000377400.8
|
CBWD1
|
COBW domain containing 1
|
|
chr10_+_94683771
Show fit
|
0.12 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18
|
|
chr3_+_195720867
Show fit
|
0.12 |
ENST00000436408.6
|
MUC20
|
mucin 20, cell surface associated
|
|
chr17_+_50746614
Show fit
|
0.11 |
ENST00000513969.5
ENST00000503728.1
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor
|
|
chr3_-_151384741
Show fit
|
0.11 |
ENST00000302632.4
|
P2RY12
|
purinergic receptor P2Y12
|
|
chr3_+_148827800
Show fit
|
0.11 |
ENST00000282957.9
ENST00000468341.1
|
CPB1
|
carboxypeptidase B1
|
|
chr8_-_86230360
Show fit
|
0.11 |
ENST00000419776.2
ENST00000297524.8
|
SLC7A13
|
solute carrier family 7 member 13
|
|
chr3_+_188153271
Show fit
|
0.11 |
ENST00000448637.5
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_140941792
Show fit
|
0.11 |
ENST00000446041.6
ENST00000324194.12
ENST00000507429.5
|
SLC25A36
|
solute carrier family 25 member 36
|
|
chr7_-_30550761
Show fit
|
0.11 |
ENST00000598361.4
ENST00000440438.6
|
ENSG00000281039.1
GARS1-DT
|
novel protein
GARS1 divergent transcript
|
|
chr20_+_36236433
Show fit
|
0.10 |
ENST00000397286.7
ENST00000679519.1
ENST00000679667.1
ENST00000680933.1
ENST00000680247.1
ENST00000320849.9
ENST00000680811.1
ENST00000373932.3
|
AAR2
|
AAR2 splicing factor
|
|
chr12_-_53499615
Show fit
|
0.10 |
ENST00000267079.6
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chr5_-_151093566
Show fit
|
0.10 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr3_-_146528750
Show fit
|
0.10 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr14_+_22543179
Show fit
|
0.09 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3
|
|
chrX_+_109535775
Show fit
|
0.09 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2
|
|
chr2_+_108621260
Show fit
|
0.09 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1
|
|
chr5_+_140786291
Show fit
|
0.09 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1
|
|
chr1_-_247078804
Show fit
|
0.09 |
ENST00000366503.3
|
ZNF670
|
zinc finger protein 670
|
|
chr12_-_10613585
Show fit
|
0.09 |
ENST00000539554.5
ENST00000625272.1
ENST00000320756.7
|
MAGOHB
|
mago homolog B, exon junction complex subunit
|
|
chr9_+_68241854
Show fit
|
0.09 |
ENST00000616550.4
ENST00000618217.4
ENST00000377342.9
ENST00000478048.5
ENST00000360171.11
|
CBWD3
|
COBW domain containing 3
|
|
chr12_+_26195313
Show fit
|
0.08 |
ENST00000422622.3
|
SSPN
|
sarcospan
|
|
chr10_-_68471911
Show fit
|
0.08 |
ENST00000358410.8
ENST00000399180.3
|
DNA2
|
DNA replication helicase/nuclease 2
|
|
chr10_-_100065394
Show fit
|
0.08 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N subunit 1
|
|
chr4_+_73853290
Show fit
|
0.08 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr7_-_32070396
Show fit
|
0.08 |
ENST00000396191.6
|
PDE1C
|
phosphodiesterase 1C
|
|
chr20_-_10420737
Show fit
|
0.08 |
ENST00000649912.1
|
ENSG00000285723.1
|
novel protein
|
|
chrX_+_85003863
Show fit
|
0.08 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like
|
|
chr15_-_43493105
Show fit
|
0.08 |
ENST00000382039.7
ENST00000450115.6
ENST00000382044.9
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr12_+_96912517
Show fit
|
0.07 |
ENST00000457368.2
|
NEDD1
|
NEDD1 gamma-tubulin ring complex targeting factor
|
|
chr3_-_119463606
Show fit
|
0.07 |
ENST00000319172.10
ENST00000491685.5
ENST00000461654.1
|
TMEM39A
|
transmembrane protein 39A
|
|
chrX_+_109536641
Show fit
|
0.07 |
ENST00000372107.5
|
NXT2
|
nuclear transport factor 2 like export factor 2
|
|
chrX_-_19670983
Show fit
|
0.07 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1
|
|
chr6_+_159800249
Show fit
|
0.07 |
ENST00000610273.5
ENST00000392167.4
|
PNLDC1
|
PARN like, ribonuclease domain containing 1
|
|
chr9_-_41189310
Show fit
|
0.07 |
ENST00000456520.5
ENST00000377391.8
ENST00000613716.4
ENST00000617933.1
|
CBWD6
|
COBW domain containing 6
|
|
chr4_-_83284752
Show fit
|
0.07 |
ENST00000311461.7
ENST00000647002.2
ENST00000311469.9
|
COQ2
|
coenzyme Q2, polyprenyltransferase
|
|
chr9_+_65675834
Show fit
|
0.07 |
ENST00000377392.9
ENST00000377384.5
ENST00000430059.6
ENST00000429800.6
ENST00000382405.8
ENST00000377395.8
|
CBWD5
|
COBW domain containing 5
|
|
chr3_-_109337572
Show fit
|
0.07 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4
|
|
chr17_+_29246852
Show fit
|
0.06 |
ENST00000225387.8
|
CRYBA1
|
crystallin beta A1
|
|
chr22_-_49825900
Show fit
|
0.06 |
ENST00000404034.5
|
BRD1
|
bromodomain containing 1
|
|
chr5_-_59216826
Show fit
|
0.06 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D
|
|
chr13_-_37059432
Show fit
|
0.06 |
ENST00000464744.5
|
SUPT20H
|
SPT20 homolog, SAGA complex component
|
|
chr10_-_68471882
Show fit
|
0.06 |
ENST00000551118.6
|
DNA2
|
DNA replication helicase/nuclease 2
|
|
chr12_-_11269696
Show fit
|
0.06 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3
|
|
chr22_+_29480211
Show fit
|
0.06 |
ENST00000310624.7
|
NEFH
|
neurofilament heavy
|
|
chr1_-_35193135
Show fit
|
0.06 |
ENST00000357214.6
|
SFPQ
|
splicing factor proline and glutamine rich
|
|
chr12_+_93677556
Show fit
|
0.06 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr19_+_41796616
Show fit
|
0.06 |
ENST00000344550.4
|
CEACAM3
|
CEA cell adhesion molecule 3
|
|
chr12_+_48472559
Show fit
|
0.05 |
ENST00000266594.4
|
ANP32D
|
acidic nuclear phosphoprotein 32 family member D
|
|
chr16_+_53703963
Show fit
|
0.05 |
ENST00000636218.1
ENST00000637001.1
ENST00000471389.6
ENST00000637969.1
ENST00000640179.1
|
FTO
|
FTO alpha-ketoglutarate dependent dioxygenase
|
|
chr14_+_39114289
Show fit
|
0.05 |
ENST00000396249.7
ENST00000250379.13
ENST00000534684.7
ENST00000308317.12
ENST00000527381.2
|
GEMIN2
|
gem nuclear organelle associated protein 2
|
|
chr10_+_133394119
Show fit
|
0.05 |
ENST00000317502.11
ENST00000432508.3
|
MTG1
|
mitochondrial ribosome associated GTPase 1
|
|
chr2_-_111884117
Show fit
|
0.05 |
ENST00000341068.8
|
ANAPC1
|
anaphase promoting complex subunit 1
|
|
chr5_+_135834641
Show fit
|
0.05 |
ENST00000425402.5
ENST00000681962.1
ENST00000650267.1
ENST00000510147.2
ENST00000274513.9
ENST00000433282.6
ENST00000412661.3
|
SLC25A48
|
solute carrier family 25 member 48
|
|
chr4_-_162163955
Show fit
|
0.05 |
ENST00000379164.8
|
FSTL5
|
follistatin like 5
|
|
chr4_-_185203923
Show fit
|
0.05 |
ENST00000296775.7
ENST00000458385.7
ENST00000514798.1
|
CFAP97
|
cilia and flagella associated protein 97
|
|
chr15_-_34754989
Show fit
|
0.05 |
ENST00000290374.5
|
GJD2
|
gap junction protein delta 2
|
|
chr15_+_78540729
Show fit
|
0.05 |
ENST00000559365.5
ENST00000558341.5
ENST00000559437.5
|
PSMA4
|
proteasome 20S subunit alpha 4
|
|
chrX_-_23907887
Show fit
|
0.05 |
ENST00000379226.9
|
APOO
|
apolipoprotein O
|
|
chr2_+_167187283
Show fit
|
0.04 |
ENST00000409605.1
ENST00000409273.6
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr13_-_46897021
Show fit
|
0.04 |
ENST00000542664.4
ENST00000543956.5
|
HTR2A
|
5-hydroxytryptamine receptor 2A
|
|
chr1_+_197268204
Show fit
|
0.04 |
ENST00000535699.5
ENST00000538660.5
|
CRB1
|
crumbs cell polarity complex component 1
|
|
chrX_+_12137409
Show fit
|
0.04 |
ENST00000672010.1
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr14_+_22304051
Show fit
|
0.04 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39
|
|
chr8_+_86342539
Show fit
|
0.03 |
ENST00000517970.6
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr19_+_21020634
Show fit
|
0.03 |
ENST00000599548.5
ENST00000594110.5
ENST00000261560.10
|
ZNF430
|
zinc finger protein 430
|
|
chr15_+_49423233
Show fit
|
0.03 |
ENST00000560270.1
ENST00000267843.9
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7
|
|
chr4_+_26343156
Show fit
|
0.03 |
ENST00000680928.1
ENST00000681260.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr2_+_167187364
Show fit
|
0.03 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr3_+_173398438
Show fit
|
0.03 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1
|
|
chr19_+_12064720
Show fit
|
0.03 |
ENST00000550826.1
ENST00000439326.8
|
ZNF844
|
zinc finger protein 844
|
|
chr3_-_24495532
Show fit
|
0.03 |
ENST00000643772.1
ENST00000642307.1
ENST00000645139.1
|
THRB
|
thyroid hormone receptor beta
|
|
chr9_+_130265745
Show fit
|
0.03 |
ENST00000683500.1
|
HMCN2
|
hemicentin 2
|
|
chr11_+_59511368
Show fit
|
0.03 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9
|
|
chr8_-_26045360
Show fit
|
0.03 |
ENST00000520164.6
|
EBF2
|
EBF transcription factor 2
|
|
chr18_+_45615473
Show fit
|
0.03 |
ENST00000255226.11
|
SLC14A2
|
solute carrier family 14 member 2
|
|
chr16_+_81739027
Show fit
|
0.03 |
ENST00000566191.5
ENST00000565272.5
ENST00000563954.5
ENST00000565054.5
|
ENSG00000261218.5
PLCG2
|
novel transcript
phospholipase C gamma 2
|
|
chr7_-_22194709
Show fit
|
0.02 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5
|
|
chr1_+_95151377
Show fit
|
0.02 |
ENST00000604203.1
|
TLCD4-RWDD3
|
TLCD4-RWDD3 readthrough
|
|
chr5_+_141100466
Show fit
|
0.02 |
ENST00000231130.3
|
PCDHB3
|
protocadherin beta 3
|
|
chr10_+_52128343
Show fit
|
0.02 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1
|
|
chr1_+_248030070
Show fit
|
0.02 |
ENST00000642011.1
ENST00000641771.1
|
OR2L2
|
olfactory receptor family 2 subfamily L member 2
|
|
chr15_-_43493076
Show fit
|
0.02 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr15_+_78540405
Show fit
|
0.02 |
ENST00000560217.5
ENST00000559082.5
ENST00000559948.5
ENST00000413382.6
ENST00000559146.5
ENST00000044462.12
ENST00000558281.5
|
PSMA4
|
proteasome 20S subunit alpha 4
|
|
chr20_-_56497608
Show fit
|
0.02 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7
|
|
chr10_+_84452208
Show fit
|
0.02 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr7_+_142791635
Show fit
|
0.02 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1
|
|
chr1_-_113871665
Show fit
|
0.02 |
ENST00000528414.5
ENST00000460620.5
ENST00000359785.10
ENST00000420377.6
ENST00000525799.1
ENST00000538253.5
|
PTPN22
|
protein tyrosine phosphatase non-receptor type 22
|
|
chr12_-_54588636
Show fit
|
0.01 |
ENST00000257905.13
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A
|
|
chr3_-_149752443
Show fit
|
0.01 |
ENST00000473414.6
|
COMMD2
|
COMM domain containing 2
|
|
chr10_+_24449426
Show fit
|
0.01 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217
|
|
chr2_-_69961624
Show fit
|
0.01 |
ENST00000320256.6
|
ASPRV1
|
aspartic peptidase retroviral like 1
|
|
chr4_-_185812209
Show fit
|
0.01 |
ENST00000393523.6
ENST00000393528.7
ENST00000449407.6
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_74391837
Show fit
|
0.01 |
ENST00000417090.1
ENST00000409868.5
ENST00000680606.1
|
DCTN1
|
dynactin subunit 1
|
|
chr1_+_160400543
Show fit
|
0.01 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2
|
|
chr7_-_128031422
Show fit
|
0.00 |
ENST00000249363.4
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr13_+_73054969
Show fit
|
0.00 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5
|
|
chr6_-_63319943
Show fit
|
0.00 |
ENST00000622415.1
ENST00000370658.9
ENST00000485906.6
ENST00000370657.9
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr12_-_11395556
Show fit
|
0.00 |
ENST00000565533.1
ENST00000389362.6
ENST00000546254.3
|
PRB2
PRB1
|
proline rich protein BstNI subfamily 2
proline rich protein BstNI subfamily 1
|
|
chr19_+_7595830
Show fit
|
0.00 |
ENST00000160298.9
ENST00000446248.4
|
CAMSAP3
|
calmodulin regulated spectrin associated protein family member 3
|
|
chr12_+_64497968
Show fit
|
0.00 |
ENST00000676593.1
ENST00000677093.1
|
TBK1
ENSG00000288665.1
|
TANK binding kinase 1
novel transcript
|
|
chr10_+_80132591
Show fit
|
0.00 |
ENST00000372267.6
|
PLAC9
|
placenta associated 9
|