Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
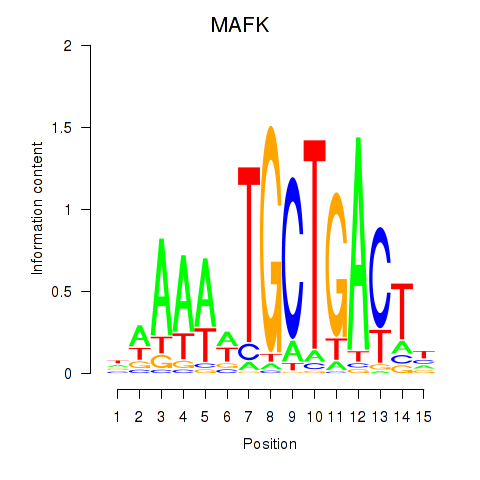
Results for MAFK
Z-value: 0.72
Transcription factors associated with MAFK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFK
|
ENSG00000198517.10 | MAFK |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFK | hg38_v1_chr7_+_1530684_1530726 | -0.43 | 1.9e-02 | Click! |
Activity profile of MAFK motif
Sorted Z-values of MAFK motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFK
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_16881967 | 4.49 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr4_+_69995958 | 3.77 |
ENST00000381060.2
ENST00000246895.9 |
STATH
|
statherin |
| chr10_-_13001705 | 1.85 |
ENST00000378825.5
|
CCDC3
|
coiled-coil domain containing 3 |
| chr11_+_62208665 | 1.55 |
ENST00000244930.6
|
SCGB2A1
|
secretoglobin family 2A member 1 |
| chr1_-_109112733 | 1.55 |
ENST00000369948.8
ENST00000369945.7 |
C1orf194
|
chromosome 1 open reading frame 194 |
| chr6_-_52803807 | 1.52 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr1_-_161367872 | 1.51 |
ENST00000367974.2
|
CFAP126
|
cilia and flagella associated protein 126 |
| chr4_-_99435396 | 1.38 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr14_-_106622837 | 1.35 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr16_+_80540980 | 1.31 |
ENST00000568035.5
ENST00000305904.11 |
DYNLRB2
|
dynein light chain roadblock-type 2 |
| chr11_+_27055215 | 1.29 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr1_-_206946448 | 1.23 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor |
| chr18_+_63702958 | 1.21 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr21_+_41322805 | 1.16 |
ENST00000398646.3
|
FAM3B
|
FAM3 metabolism regulating signaling molecule B |
| chr13_-_38990856 | 1.16 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr3_+_13568721 | 1.14 |
ENST00000492059.5
|
FBLN2
|
fibulin 2 |
| chr19_-_8896090 | 1.13 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr10_-_96358989 | 1.10 |
ENST00000371172.8
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chrY_+_2841594 | 1.01 |
ENST00000250784.13
|
RPS4Y1
|
ribosomal protein S4 Y-linked 1 |
| chr12_+_110614097 | 1.00 |
ENST00000471804.7
ENST00000377654.5 ENST00000614115.5 ENST00000397655.7 ENST00000549123.6 ENST00000679401.1 ENST00000397659.9 ENST00000680445.1 |
TCTN1
|
tectonic family member 1 |
| chr5_-_35938572 | 0.98 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr6_+_32844789 | 0.97 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr4_-_99435134 | 0.97 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_+_171522227 | 0.96 |
ENST00000409484.5
|
CYBRD1
|
cytochrome b reductase 1 |
| chr2_+_8682046 | 0.94 |
ENST00000331129.3
ENST00000396290.2 |
ID2
|
inhibitor of DNA binding 2 |
| chr14_-_106185387 | 0.93 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr4_-_69860138 | 0.93 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr10_-_114684457 | 0.93 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr13_+_24160740 | 0.93 |
ENST00000382095.8
|
SPATA13
|
spermatogenesis associated 13 |
| chr12_-_10130241 | 0.91 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr5_-_16508788 | 0.88 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr12_-_10130082 | 0.87 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chrY_+_20575792 | 0.86 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chr15_-_56465130 | 0.86 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chr12_-_10130143 | 0.84 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr8_-_109648825 | 0.81 |
ENST00000533895.5
ENST00000446070.6 ENST00000528331.5 ENST00000526302.5 ENST00000408908.6 ENST00000433638.1 ENST00000524720.5 |
SYBU
|
syntabulin |
| chr4_-_69653223 | 0.81 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr12_+_110614027 | 0.78 |
ENST00000550703.6
ENST00000551590.5 |
TCTN1
|
tectonic family member 1 |
| chr1_-_89126066 | 0.78 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr3_-_94062906 | 0.78 |
ENST00000314636.3
ENST00000394221.3 |
DHFR2
|
dihydrofolate reductase 2 |
| chr6_+_33075952 | 0.76 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chrY_+_20575716 | 0.74 |
ENST00000361365.7
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chr3_+_42906107 | 0.73 |
ENST00000440367.7
ENST00000328199.6 |
ZNF662
|
zinc finger protein 662 |
| chr18_-_27143024 | 0.73 |
ENST00000581714.5
|
CHST9
|
carbohydrate sulfotransferase 9 |
| chr3_-_167407837 | 0.72 |
ENST00000455345.7
|
ZBBX
|
zinc finger B-box domain containing |
| chr4_+_95051671 | 0.71 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr13_+_36819214 | 0.71 |
ENST00000255476.3
|
RFXAP
|
regulatory factor X associated protein |
| chr12_-_68302872 | 0.70 |
ENST00000539972.5
|
MDM1
|
Mdm1 nuclear protein |
| chr16_+_19410480 | 0.70 |
ENST00000541464.5
|
TMC5
|
transmembrane channel like 5 |
| chr16_+_19410723 | 0.69 |
ENST00000381414.8
ENST00000396229.6 |
TMC5
|
transmembrane channel like 5 |
| chr5_+_140875299 | 0.68 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chrX_+_35798342 | 0.67 |
ENST00000399988.5
ENST00000399992.5 ENST00000399987.5 ENST00000399989.5 |
MAGEB16
|
MAGE family member B16 |
| chr11_-_26567087 | 0.66 |
ENST00000436318.6
ENST00000281268.12 |
MUC15
|
mucin 15, cell surface associated |
| chr7_-_138627444 | 0.66 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr17_+_9825906 | 0.66 |
ENST00000262441.10
|
GLP2R
|
glucagon like peptide 2 receptor |
| chr5_+_140834230 | 0.64 |
ENST00000356878.5
ENST00000525929.2 |
PCDHA7
|
protocadherin alpha 7 |
| chr6_+_131637296 | 0.63 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_-_150765785 | 0.62 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr1_-_150765735 | 0.62 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr8_+_109086585 | 0.61 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr12_-_91180365 | 0.59 |
ENST00000547937.5
|
DCN
|
decorin |
| chr5_+_157269317 | 0.58 |
ENST00000618329.4
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_-_68206039 | 0.58 |
ENST00000395463.3
|
CALML4
|
calmodulin like 4 |
| chr6_+_159800249 | 0.58 |
ENST00000610273.5
ENST00000392167.4 |
PNLDC1
|
PARN like, ribonuclease domain containing 1 |
| chr16_+_82035245 | 0.58 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr8_-_109680812 | 0.57 |
ENST00000528716.5
ENST00000527600.5 ENST00000531230.5 ENST00000532189.5 ENST00000534184.5 ENST00000408889.7 ENST00000533171.5 |
SYBU
|
syntabulin |
| chr8_-_132625378 | 0.57 |
ENST00000522789.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr1_+_85062304 | 0.57 |
ENST00000326813.12
ENST00000528899.5 ENST00000294664.11 |
DNAI3
|
dynein axonemal intermediate chain 3 |
| chr6_+_29439939 | 0.57 |
ENST00000622521.1
|
OR10C1
|
olfactory receptor family 10 subfamily C member 1 |
| chr6_-_52763473 | 0.56 |
ENST00000493422.3
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr1_+_183805105 | 0.56 |
ENST00000360851.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr3_-_94028582 | 0.56 |
ENST00000315099.3
|
STX19
|
syntaxin 19 |
| chr7_+_77122609 | 0.56 |
ENST00000285871.5
|
CCDC146
|
coiled-coil domain containing 146 |
| chr5_+_140827950 | 0.56 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr15_-_55365231 | 0.55 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr12_+_20815672 | 0.55 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr4_+_112297341 | 0.54 |
ENST00000504176.6
ENST00000650871.1 ENST00000177648.13 ENST00000458497.6 |
ALPK1
|
alpha kinase 1 |
| chr10_-_114684612 | 0.53 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr7_+_12570712 | 0.53 |
ENST00000417018.1
ENST00000297029.10 |
SCIN
|
scinderin |
| chr13_-_38990824 | 0.53 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr1_+_244970354 | 0.52 |
ENST00000366521.7
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr16_-_75556214 | 0.52 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr5_+_140841183 | 0.52 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr12_+_69348372 | 0.51 |
ENST00000261267.7
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr5_-_16508812 | 0.50 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr9_-_41128681 | 0.49 |
ENST00000622588.2
|
FOXD4L6
|
forkhead box D4 like 6 |
| chr8_+_27491125 | 0.48 |
ENST00000517536.5
ENST00000521400.6 ENST00000521780.5 ENST00000380476.7 ENST00000518379.5 ENST00000521684.1 |
EPHX2
|
epoxide hydrolase 2 |
| chr20_-_56525925 | 0.47 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr1_-_223665685 | 0.47 |
ENST00000366872.10
|
CAPN8
|
calpain 8 |
| chr12_-_113335030 | 0.46 |
ENST00000552014.5
ENST00000680972.1 ENST00000548186.5 ENST00000202831.7 ENST00000549181.5 |
SLC8B1
|
solute carrier family 8 member B1 |
| chr18_-_55351977 | 0.46 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr1_-_206921867 | 0.46 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr11_+_101914997 | 0.46 |
ENST00000263468.13
|
CEP126
|
centrosomal protein 126 |
| chr3_-_121660892 | 0.46 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_89526164 | 0.45 |
ENST00000548729.5
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr9_+_96928310 | 0.45 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr5_+_55102635 | 0.45 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr1_-_169734064 | 0.45 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr4_+_69096467 | 0.44 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr4_-_109801978 | 0.44 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr6_+_158649997 | 0.44 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr5_+_42548043 | 0.44 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr16_+_75222609 | 0.44 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr1_-_147225568 | 0.43 |
ENST00000533174.5
ENST00000578284.5 |
FMO5
|
flavin containing dimethylaniline monoxygenase 5 |
| chr16_+_30199860 | 0.43 |
ENST00000395138.6
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr1_-_206921987 | 0.43 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr9_+_105700953 | 0.43 |
ENST00000374688.5
|
TMEM38B
|
transmembrane protein 38B |
| chr9_+_101185029 | 0.43 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr1_-_108200849 | 0.42 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr10_-_25062279 | 0.42 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr22_+_23145366 | 0.42 |
ENST00000341989.9
ENST00000263116.8 |
RAB36
|
RAB36, member RAS oncogene family |
| chr11_+_36375978 | 0.41 |
ENST00000378867.7
|
PRR5L
|
proline rich 5 like |
| chr16_+_19067989 | 0.41 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7, hydroxylase |
| chr12_-_11022620 | 0.41 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr1_+_103617427 | 0.41 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr13_+_76992412 | 0.41 |
ENST00000636705.1
|
CLN5
|
CLN5 intracellular trafficking protein |
| chr10_-_73591330 | 0.41 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr5_-_16616972 | 0.40 |
ENST00000682564.1
ENST00000306320.10 ENST00000682229.1 |
RETREG1
|
reticulophagy regulator 1 |
| chr15_+_62561361 | 0.40 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr11_-_26572254 | 0.40 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr5_-_16508990 | 0.40 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chr18_-_63661884 | 0.39 |
ENST00000332821.8
ENST00000283752.10 |
SERPINB3
|
serpin family B member 3 |
| chr1_+_18480930 | 0.39 |
ENST00000400664.3
|
KLHDC7A
|
kelch domain containing 7A |
| chr6_-_49744434 | 0.39 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr7_-_138755892 | 0.39 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr11_+_72080313 | 0.39 |
ENST00000307198.11
ENST00000538413.6 ENST00000642648.1 ENST00000289488.7 |
ENSG00000284922.2
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr12_-_89526253 | 0.38 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr11_-_5227063 | 0.38 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr11_+_36296281 | 0.38 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr5_+_43602648 | 0.38 |
ENST00000505678.6
ENST00000512422.5 ENST00000264663.9 ENST00000670904.1 ENST00000653251.1 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr6_-_134318097 | 0.38 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr22_+_35383106 | 0.37 |
ENST00000678411.1
|
HMOX1
|
heme oxygenase 1 |
| chr15_-_89815332 | 0.37 |
ENST00000559874.2
|
ANPEP
|
alanyl aminopeptidase, membrane |
| chr6_-_98947911 | 0.37 |
ENST00000369244.7
ENST00000229971.2 |
FBXL4
|
F-box and leucine rich repeat protein 4 |
| chr14_+_22226711 | 0.37 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr15_+_73926443 | 0.36 |
ENST00000261921.8
|
LOXL1
|
lysyl oxidase like 1 |
| chr20_+_2295994 | 0.36 |
ENST00000381458.6
|
TGM3
|
transglutaminase 3 |
| chr4_-_103099811 | 0.36 |
ENST00000504285.5
ENST00000296424.9 |
BDH2
|
3-hydroxybutyrate dehydrogenase 2 |
| chr6_+_167291329 | 0.36 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A |
| chr22_+_38952718 | 0.35 |
ENST00000402255.5
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3A |
| chr6_+_46793379 | 0.35 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr13_+_73054969 | 0.35 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5 |
| chr6_+_167291309 | 0.35 |
ENST00000230256.8
|
UNC93A
|
unc-93 homolog A |
| chr4_-_87391149 | 0.35 |
ENST00000507286.1
ENST00000358290.9 |
HSD17B11
|
hydroxysteroid 17-beta dehydrogenase 11 |
| chr17_-_69268812 | 0.34 |
ENST00000586811.1
|
ABCA5
|
ATP binding cassette subfamily A member 5 |
| chr14_-_95714114 | 0.34 |
ENST00000402399.6
ENST00000555202.1 |
TCL1A
|
TCL1 family AKT coactivator A |
| chr4_-_139302460 | 0.34 |
ENST00000394223.2
ENST00000676245.1 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr3_+_113947901 | 0.34 |
ENST00000330212.7
ENST00000498275.5 |
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23 |
| chr3_-_112975018 | 0.34 |
ENST00000471858.5
ENST00000308611.8 ENST00000295863.4 |
CD200R1
|
CD200 receptor 1 |
| chr8_+_2045037 | 0.34 |
ENST00000262113.9
|
MYOM2
|
myomesin 2 |
| chr19_+_15949008 | 0.33 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr4_-_122922442 | 0.33 |
ENST00000510735.1
ENST00000304430.10 |
NUDT6
|
nudix hydrolase 6 |
| chr8_+_2045058 | 0.33 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr18_-_28036585 | 0.33 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr13_+_50015438 | 0.33 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr17_-_19745602 | 0.33 |
ENST00000444455.5
ENST00000439102.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr15_-_89655428 | 0.33 |
ENST00000394412.8
|
KIF7
|
kinesin family member 7 |
| chr11_-_34511710 | 0.33 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr11_+_26994102 | 0.32 |
ENST00000318627.4
|
FIBIN
|
fin bud initiation factor homolog |
| chr19_-_39934626 | 0.32 |
ENST00000616721.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr19_-_39245006 | 0.32 |
ENST00000413851.3
ENST00000613087.4 |
IFNL3
|
interferon lambda 3 |
| chr12_-_51009264 | 0.32 |
ENST00000545993.7
|
SLC11A2
|
solute carrier family 11 member 2 |
| chr13_-_85799400 | 0.32 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr6_-_47042306 | 0.32 |
ENST00000371253.7
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr6_+_6588082 | 0.31 |
ENST00000379953.6
|
LY86
|
lymphocyte antigen 86 |
| chr5_-_16508858 | 0.31 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr14_-_95714088 | 0.31 |
ENST00000556450.5
|
TCL1A
|
TCL1 family AKT coactivator A |
| chr1_-_162376841 | 0.31 |
ENST00000367935.10
|
SPATA46
|
spermatogenesis associated 46 |
| chr6_-_49744378 | 0.31 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr14_-_95714146 | 0.31 |
ENST00000554012.5
|
TCL1A
|
TCL1 family AKT coactivator A |
| chr5_+_90899183 | 0.31 |
ENST00000640815.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr5_-_16508951 | 0.30 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_-_182589239 | 0.30 |
ENST00000367559.7
ENST00000539397.1 |
RNASEL
|
ribonuclease L |
| chr2_-_177552781 | 0.30 |
ENST00000408939.4
|
TTC30B
|
tetratricopeptide repeat domain 30B |
| chr10_+_71319249 | 0.30 |
ENST00000373189.6
ENST00000479577.2 |
SLC29A3
|
solute carrier family 29 member 3 |
| chr6_-_28252246 | 0.30 |
ENST00000377294.3
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr1_+_3690654 | 0.30 |
ENST00000378285.5
ENST00000378280.5 ENST00000378288.8 |
TP73
|
tumor protein p73 |
| chrX_+_71118515 | 0.30 |
ENST00000333646.10
|
MED12
|
mediator complex subunit 12 |
| chr9_-_5833014 | 0.30 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr12_-_85836372 | 0.30 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr13_-_99258366 | 0.30 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
| chr16_+_19067893 | 0.30 |
ENST00000544894.6
ENST00000561858.5 |
COQ7
|
coenzyme Q7, hydroxylase |
| chr17_+_17972813 | 0.30 |
ENST00000582416.5
ENST00000313838.12 ENST00000399187.6 ENST00000581264.5 ENST00000479684.2 ENST00000584166.5 ENST00000585108.5 ENST00000399182.5 ENST00000579977.1 |
DRC3
|
dynein regulatory complex subunit 3 |
| chr16_-_28623330 | 0.30 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr15_+_70936487 | 0.29 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_+_73110375 | 0.29 |
ENST00000617744.4
ENST00000544879.5 ENST00000537969.5 ENST00000357321.9 ENST00000349051.9 |
NUDT13
|
nudix hydrolase 13 |
| chr16_-_67936808 | 0.29 |
ENST00000358514.9
|
PSMB10
|
proteasome 20S subunit beta 10 |
| chr11_+_5351508 | 0.29 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chr6_+_125919296 | 0.29 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_27247690 | 0.28 |
ENST00000421826.6
ENST00000230582.8 |
PRSS16
|
serine protease 16 |
| chr4_+_165378998 | 0.28 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E |
| chr9_+_68302867 | 0.28 |
ENST00000342833.4
|
FOXD4L3
|
forkhead box D4 like 3 |
| chr6_+_32741382 | 0.28 |
ENST00000374940.4
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr6_-_25042003 | 0.28 |
ENST00000510784.8
|
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr6_-_79234619 | 0.28 |
ENST00000344726.9
ENST00000275036.11 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr6_-_79234713 | 0.27 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr17_-_19745369 | 0.27 |
ENST00000573368.5
ENST00000457500.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr3_-_112974912 | 0.27 |
ENST00000440122.6
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr9_+_83980757 | 0.27 |
ENST00000325875.7
ENST00000445877.6 |
RMI1
|
RecQ mediated genome instability 1 |
| chr14_-_25050111 | 0.27 |
ENST00000323944.9
|
STXBP6
|
syntaxin binding protein 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.4 | 1.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.4 | 1.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 2.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 0.9 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.3 | 0.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.3 | 3.8 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 0.6 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 0.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 0.5 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 1.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 2.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.4 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.6 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.4 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 1.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.9 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.2 | GO:2001302 | regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.3 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.6 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 2.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.8 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.1 | 0.2 | GO:0100009 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 1.6 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.2 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.2 | GO:0060903 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.3 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0036046 | protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) peptidyl-lysine deglutarylation(GO:0061699) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 1.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.3 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.4 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.0 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 2.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.3 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 2.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 1.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) psychomotor behavior(GO:0036343) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 1.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.0 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 2.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 3.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.6 | 2.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 1.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.9 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 1.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 4.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.7 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.4 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 2.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0036055 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.0 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0052840 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.0 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |