Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
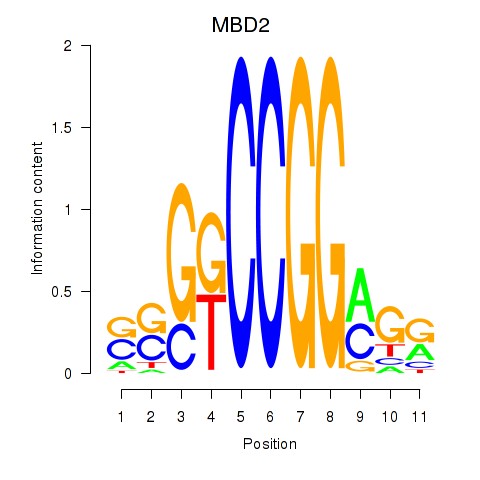
Results for MBD2
Z-value: 0.71
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.12 | MBD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg38_v1_chr18_-_54224578_54224689 | -0.24 | 2.1e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_22143088 | 1.61 |
ENST00000290167.11
|
WNT4
|
Wnt family member 4 |
| chr17_-_181640 | 0.93 |
ENST00000613549.3
|
DOC2B
|
double C2 domain beta |
| chr5_+_77210667 | 0.83 |
ENST00000264917.10
|
PDE8B
|
phosphodiesterase 8B |
| chr5_+_77210881 | 0.83 |
ENST00000340978.7
ENST00000346042.7 ENST00000342343.8 ENST00000333194.8 |
PDE8B
|
phosphodiesterase 8B |
| chr4_-_25862979 | 0.77 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3 |
| chr13_+_98143410 | 0.69 |
ENST00000596580.2
ENST00000376581.9 |
FARP1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr10_-_121598396 | 0.65 |
ENST00000336553.10
ENST00000457416.6 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr10_-_121598359 | 0.63 |
ENST00000684153.1
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr7_-_51316754 | 0.63 |
ENST00000632460.1
ENST00000441453.5 ENST00000648294.1 ENST00000265136.12 ENST00000395542.6 ENST00000395540.6 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr10_-_121598412 | 0.63 |
ENST00000360144.7
ENST00000358487.10 ENST00000369059.5 ENST00000613048.4 ENST00000356226.8 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr2_+_42048012 | 0.59 |
ENST00000294964.6
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr17_-_65561137 | 0.58 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr11_-_75525925 | 0.56 |
ENST00000336898.8
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr22_+_39456996 | 0.56 |
ENST00000341184.7
|
MGAT3
|
beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase |
| chr1_-_160262522 | 0.55 |
ENST00000440682.5
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_-_160262501 | 0.54 |
ENST00000447377.5
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr8_-_139702998 | 0.52 |
ENST00000303015.2
|
KCNK9
|
potassium two pore domain channel subfamily K member 9 |
| chr14_+_52552830 | 0.50 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr19_+_8209300 | 0.47 |
ENST00000558268.5
ENST00000558331.5 |
CERS4
|
ceramide synthase 4 |
| chr14_-_89417148 | 0.47 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chr8_+_135457442 | 0.46 |
ENST00000355849.10
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr8_+_135458027 | 0.46 |
ENST00000524199.5
|
KHDRBS3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr9_+_36136703 | 0.46 |
ENST00000377960.9
ENST00000377959.5 |
GLIPR2
|
GLI pathogenesis related 2 |
| chr16_+_28292485 | 0.46 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr15_-_45187955 | 0.45 |
ENST00000560471.5
ENST00000560540.5 |
SHF
|
Src homology 2 domain containing F |
| chr5_+_10564064 | 0.45 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr19_-_18606779 | 0.44 |
ENST00000684169.1
ENST00000392386.8 |
CRLF1
|
cytokine receptor like factor 1 |
| chr9_+_36136752 | 0.43 |
ENST00000619700.1
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr22_-_19431692 | 0.43 |
ENST00000340170.8
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr16_+_2969307 | 0.42 |
ENST00000576565.1
ENST00000318782.9 |
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr17_+_67825664 | 0.42 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor |
| chr16_+_2969270 | 0.42 |
ENST00000293978.12
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr9_+_36136416 | 0.42 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr16_+_2969548 | 0.41 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr11_+_2444986 | 0.41 |
ENST00000155840.12
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chrX_+_153072454 | 0.41 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A |
| chr19_+_8209320 | 0.40 |
ENST00000561053.5
ENST00000559450.5 ENST00000251363.10 ENST00000559336.5 |
CERS4
|
ceramide synthase 4 |
| chr4_-_16226460 | 0.39 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr15_-_45187947 | 0.39 |
ENST00000560734.5
|
SHF
|
Src homology 2 domain containing F |
| chr3_+_184561768 | 0.38 |
ENST00000330394.3
|
EPHB3
|
EPH receptor B3 |
| chr1_+_43389874 | 0.38 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr9_-_134068012 | 0.38 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr22_-_31346317 | 0.37 |
ENST00000266269.10
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr19_-_461007 | 0.37 |
ENST00000264554.11
|
SHC2
|
SHC adaptor protein 2 |
| chr10_+_115093404 | 0.37 |
ENST00000527407.5
|
ATRNL1
|
attractin like 1 |
| chr22_-_31346143 | 0.36 |
ENST00000405309.7
ENST00000351933.8 |
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr13_+_34942263 | 0.36 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr18_-_55588535 | 0.36 |
ENST00000566286.5
ENST00000566279.5 ENST00000626595.2 ENST00000564999.5 ENST00000616053.4 ENST00000356073.8 |
TCF4
|
transcription factor 4 |
| chr12_+_45729899 | 0.36 |
ENST00000422737.6
|
ARID2
|
AT-rich interaction domain 2 |
| chr16_+_58025745 | 0.36 |
ENST00000219271.4
|
MMP15
|
matrix metallopeptidase 15 |
| chr5_+_128083757 | 0.35 |
ENST00000262461.7
ENST00000628403.2 ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 member 2 |
| chr10_-_121598234 | 0.35 |
ENST00000369058.7
ENST00000369060.8 ENST00000359354.6 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr2_+_235494024 | 0.35 |
ENST00000304032.13
ENST00000409457.5 ENST00000336665.9 |
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr18_-_55588184 | 0.35 |
ENST00000354452.8
ENST00000565908.6 ENST00000635822.2 |
TCF4
|
transcription factor 4 |
| chr17_+_38705482 | 0.34 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chr4_+_4387078 | 0.34 |
ENST00000504171.1
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr7_+_129188622 | 0.34 |
ENST00000249373.8
|
SMO
|
smoothened, frizzled class receptor |
| chr11_+_43358908 | 0.33 |
ENST00000039989.9
ENST00000299240.10 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr1_+_87328860 | 0.33 |
ENST00000370544.10
|
LMO4
|
LIM domain only 4 |
| chr2_+_216633411 | 0.33 |
ENST00000233809.9
|
IGFBP2
|
insulin like growth factor binding protein 2 |
| chr17_+_59331633 | 0.32 |
ENST00000312655.9
|
YPEL2
|
yippee like 2 |
| chr1_+_6785437 | 0.31 |
ENST00000303635.12
ENST00000473578.5 ENST00000557126.5 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr17_+_50560703 | 0.31 |
ENST00000359106.10
|
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr1_-_54406385 | 0.31 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr9_-_109498251 | 0.30 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr19_+_10416773 | 0.30 |
ENST00000592685.5
|
PDE4A
|
phosphodiesterase 4A |
| chr14_+_52553273 | 0.30 |
ENST00000542169.6
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr3_-_187745460 | 0.30 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr3_-_18425295 | 0.30 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr2_+_127701490 | 0.30 |
ENST00000310981.6
|
SFT2D3
|
SFT2 domain containing 3 |
| chr7_-_150978284 | 0.30 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr3_-_36945042 | 0.29 |
ENST00000645898.2
ENST00000429976.5 |
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr10_-_102120246 | 0.29 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chr8_-_141308280 | 0.28 |
ENST00000517878.6
|
SLC45A4
|
solute carrier family 45 member 4 |
| chr3_+_113211459 | 0.28 |
ENST00000495514.5
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr17_+_67825494 | 0.28 |
ENST00000306378.11
ENST00000544778.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr6_-_46015812 | 0.28 |
ENST00000544153.3
ENST00000339561.12 |
CLIC5
|
chloride intracellular channel 5 |
| chr15_-_65422894 | 0.28 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4 |
| chr3_+_113211539 | 0.28 |
ENST00000682979.1
ENST00000485230.5 |
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr4_+_1793285 | 0.27 |
ENST00000440486.8
ENST00000412135.7 ENST00000481110.7 ENST00000340107.8 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr2_-_25341886 | 0.27 |
ENST00000321117.10
|
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr17_-_28951285 | 0.26 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12 |
| chr18_+_79395856 | 0.26 |
ENST00000253506.9
ENST00000591814.5 ENST00000427363.7 |
NFATC1
|
nuclear factor of activated T cells 1 |
| chr17_+_50561010 | 0.26 |
ENST00000360761.8
ENST00000354983.8 ENST00000352832.9 |
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr2_+_218672291 | 0.26 |
ENST00000440309.5
ENST00000424080.1 |
STK36
|
serine/threonine kinase 36 |
| chr21_-_41953997 | 0.26 |
ENST00000380486.4
|
C2CD2
|
C2 calcium dependent domain containing 2 |
| chr16_+_57735723 | 0.25 |
ENST00000562592.5
ENST00000379661.8 ENST00000566726.5 |
KATNB1
|
katanin regulatory subunit B1 |
| chr20_-_610299 | 0.25 |
ENST00000246080.4
|
TCF15
|
transcription factor 15 |
| chr1_-_111989608 | 0.25 |
ENST00000302127.5
|
KCND3
|
potassium voltage-gated channel subfamily D member 3 |
| chrX_-_137033991 | 0.25 |
ENST00000651716.2
|
GPR101
|
G protein-coupled receptor 101 |
| chr22_+_28883564 | 0.25 |
ENST00000544604.7
|
ZNRF3
|
zinc and ring finger 3 |
| chr20_+_37903104 | 0.25 |
ENST00000373459.4
ENST00000373461.9 ENST00000448944.1 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr19_+_49119531 | 0.24 |
ENST00000334186.9
|
PPFIA3
|
PTPRF interacting protein alpha 3 |
| chr1_-_17439657 | 0.24 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr5_-_180649613 | 0.24 |
ENST00000393347.7
ENST00000619105.4 |
FLT4
|
fms related receptor tyrosine kinase 4 |
| chr6_-_3457018 | 0.24 |
ENST00000436008.6
ENST00000406686.8 |
SLC22A23
|
solute carrier family 22 member 23 |
| chr9_-_124771304 | 0.23 |
ENST00000416460.6
ENST00000487099.7 |
NR6A1
|
nuclear receptor subfamily 6 group A member 1 |
| chr4_+_4387039 | 0.23 |
ENST00000621129.4
|
NSG1
|
neuronal vesicle trafficking associated 1 |
| chr9_+_22446808 | 0.23 |
ENST00000325870.3
|
DMRTA1
|
DMRT like family A1 |
| chr1_-_32336224 | 0.23 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1 |
| chr10_-_102120318 | 0.23 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1 |
| chr9_-_86099506 | 0.22 |
ENST00000388712.7
|
GOLM1
|
golgi membrane protein 1 |
| chr5_+_123512087 | 0.22 |
ENST00000345990.8
|
CSNK1G3
|
casein kinase 1 gamma 3 |
| chr22_-_17121311 | 0.22 |
ENST00000331437.4
ENST00000399875.1 |
TMEM121B
|
transmembrane protein 121B |
| chr14_-_39432414 | 0.22 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr17_+_38705243 | 0.21 |
ENST00000621332.5
|
MLLT6
|
MLLT6, PHD finger containing |
| chr1_-_50423431 | 0.21 |
ENST00000404795.4
|
DMRTA2
|
DMRT like family A2 |
| chr5_-_180071708 | 0.21 |
ENST00000522208.6
ENST00000521389.6 |
RNF130
|
ring finger protein 130 |
| chr17_+_66964638 | 0.21 |
ENST00000262138.4
|
CACNG4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr9_-_3525968 | 0.21 |
ENST00000382004.7
ENST00000617270.5 ENST00000449190.5 |
RFX3
|
regulatory factor X3 |
| chr17_+_19648792 | 0.21 |
ENST00000630662.2
|
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr2_-_213151590 | 0.21 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr2_+_218672027 | 0.21 |
ENST00000392105.7
ENST00000455724.5 ENST00000295709.8 |
STK36
|
serine/threonine kinase 36 |
| chr15_-_73753308 | 0.21 |
ENST00000569673.3
|
INSYN1
|
inhibitory synaptic factor 1 |
| chr20_+_58692767 | 0.21 |
ENST00000356091.11
|
NPEPL1
|
aminopeptidase like 1 |
| chr11_-_64917200 | 0.20 |
ENST00000377264.8
ENST00000421419.3 |
ATG2A
|
autophagy related 2A |
| chr11_-_45665578 | 0.20 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1 |
| chr17_+_29593468 | 0.20 |
ENST00000614878.4
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr19_+_5805018 | 0.20 |
ENST00000303212.3
|
NRTN
|
neurturin |
| chr22_-_20016807 | 0.19 |
ENST00000263207.8
|
ARVCF
|
ARVCF delta catenin family member |
| chr17_-_44199206 | 0.19 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3 |
| chr14_+_102362931 | 0.19 |
ENST00000359520.12
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr3_+_141402322 | 0.19 |
ENST00000510338.5
ENST00000504673.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr20_-_44810539 | 0.19 |
ENST00000372851.8
|
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr7_+_73667824 | 0.19 |
ENST00000324941.5
ENST00000451519.1 |
VPS37D
|
VPS37D subunit of ESCRT-I |
| chr1_+_33256479 | 0.19 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr3_-_196568575 | 0.19 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr19_+_3366549 | 0.19 |
ENST00000341919.7
ENST00000590282.5 ENST00000443272.3 |
NFIC
|
nuclear factor I C |
| chr10_+_114043858 | 0.19 |
ENST00000369295.4
|
ADRB1
|
adrenoceptor beta 1 |
| chr8_-_22131003 | 0.19 |
ENST00000381418.9
|
HR
|
HR lysine demethylase and nuclear receptor corepressor |
| chr2_-_37672178 | 0.18 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr12_-_54588636 | 0.18 |
ENST00000257905.13
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A |
| chr17_+_19648723 | 0.18 |
ENST00000672357.1
ENST00000584332.6 ENST00000176643.11 ENST00000339618.8 ENST00000579855.5 ENST00000671878.1 ENST00000395575.7 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr1_-_154558650 | 0.18 |
ENST00000292211.5
|
UBE2Q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr8_+_98426943 | 0.18 |
ENST00000287042.5
|
KCNS2
|
potassium voltage-gated channel modifier subfamily S member 2 |
| chr12_-_49828394 | 0.18 |
ENST00000335999.7
|
NCKAP5L
|
NCK associated protein 5 like |
| chr3_-_196568596 | 0.18 |
ENST00000433160.1
|
WDR53
|
WD repeat domain 53 |
| chr1_-_31764035 | 0.18 |
ENST00000373655.6
|
ADGRB2
|
adhesion G protein-coupled receptor B2 |
| chr11_+_119107335 | 0.18 |
ENST00000648610.2
ENST00000336702.7 |
C2CD2L
|
C2CD2 like |
| chr11_+_100687279 | 0.18 |
ENST00000298815.13
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr4_+_1793776 | 0.18 |
ENST00000352904.6
|
FGFR3
|
fibroblast growth factor receptor 3 |
| chr17_+_19648915 | 0.18 |
ENST00000672567.1
ENST00000672709.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr11_+_123430259 | 0.18 |
ENST00000533341.3
ENST00000635736.2 |
GRAMD1B
|
GRAM domain containing 1B |
| chr17_-_6556447 | 0.18 |
ENST00000421306.7
|
PITPNM3
|
PITPNM family member 3 |
| chr16_-_2340703 | 0.18 |
ENST00000301732.10
ENST00000382381.7 |
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr22_-_45977154 | 0.18 |
ENST00000339464.9
|
WNT7B
|
Wnt family member 7B |
| chr1_-_31763866 | 0.18 |
ENST00000398547.5
|
ADGRB2
|
adhesion G protein-coupled receptor B2 |
| chrX_+_69504320 | 0.17 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B |
| chr12_+_119989869 | 0.17 |
ENST00000397558.6
|
BICDL1
|
BICD family like cargo adaptor 1 |
| chr15_+_91853819 | 0.17 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family member 3A1 |
| chr17_+_40342342 | 0.17 |
ENST00000394081.7
|
RARA
|
retinoic acid receptor alpha |
| chr11_-_1572261 | 0.17 |
ENST00000397374.8
|
DUSP8
|
dual specificity phosphatase 8 |
| chr8_-_143160248 | 0.17 |
ENST00000342752.9
|
LY6H
|
lymphocyte antigen 6 family member H |
| chr6_+_125154189 | 0.17 |
ENST00000532429.5
ENST00000534199.5 |
TPD52L1
|
TPD52 like 1 |
| chr17_-_63700100 | 0.17 |
ENST00000578993.5
ENST00000259006.8 ENST00000583211.5 |
LIMD2
|
LIM domain containing 2 |
| chr17_-_28951443 | 0.17 |
ENST00000268756.7
ENST00000332830.9 ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr11_-_59615673 | 0.17 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein |
| chr10_+_126905409 | 0.17 |
ENST00000280333.9
ENST00000623213.2 |
DOCK1
|
dedicator of cytokinesis 1 |
| chr6_-_93419545 | 0.17 |
ENST00000369297.1
ENST00000369303.9 ENST00000680224.1 ENST00000681532.1 ENST00000679565.1 |
EPHA7
|
EPH receptor A7 |
| chr12_-_47819866 | 0.17 |
ENST00000354334.7
ENST00000430670.5 ENST00000552960.5 ENST00000440293.5 ENST00000080059.12 |
HDAC7
|
histone deacetylase 7 |
| chr18_-_79988526 | 0.17 |
ENST00000591711.5
ENST00000588162.1 ENST00000269601.10 |
TXNL4A
|
thioredoxin like 4A |
| chr10_-_124744280 | 0.16 |
ENST00000337318.8
|
FAM53B
|
family with sequence similarity 53 member B |
| chr12_-_49060742 | 0.16 |
ENST00000301067.12
ENST00000683543.1 |
KMT2D
|
lysine methyltransferase 2D |
| chr17_-_7238171 | 0.16 |
ENST00000574236.5
ENST00000572789.5 |
PHF23
|
PHD finger protein 23 |
| chr20_+_35542038 | 0.16 |
ENST00000357394.8
ENST00000348547.7 ENST00000416206.5 ENST00000640748.1 ENST00000411577.5 ENST00000413587.5 |
ERGIC3
|
ERGIC and golgi 3 |
| chr1_+_156369202 | 0.16 |
ENST00000537040.6
|
RHBG
|
Rh family B glycoprotein |
| chr1_+_225810057 | 0.16 |
ENST00000272167.10
ENST00000448202.5 |
EPHX1
|
epoxide hydrolase 1 |
| chr3_-_196568542 | 0.16 |
ENST00000332629.7
|
WDR53
|
WD repeat domain 53 |
| chr7_-_127392310 | 0.16 |
ENST00000393312.5
|
ZNF800
|
zinc finger protein 800 |
| chr18_+_74597850 | 0.16 |
ENST00000582337.5
ENST00000299687.10 |
ZNF407
|
zinc finger protein 407 |
| chr11_+_65084257 | 0.16 |
ENST00000526791.1
ENST00000526945.5 |
ZFPL1
|
zinc finger protein like 1 |
| chr17_+_75525682 | 0.16 |
ENST00000392550.8
ENST00000167462.9 ENST00000375227.8 ENST00000578363.5 ENST00000579392.5 |
LLGL2
|
LLGL scribble cell polarity complex component 2 |
| chrX_-_53321319 | 0.16 |
ENST00000640694.1
ENST00000674510.1 ENST00000675719.1 ENST00000642864.1 |
IQSEC2
|
IQ motif and Sec7 domain ArfGEF 2 |
| chr8_-_144462848 | 0.15 |
ENST00000530374.6
|
CYHR1
|
cysteine and histidine rich 1 |
| chr13_+_73058993 | 0.15 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr11_-_31817904 | 0.15 |
ENST00000423822.7
|
PAX6
|
paired box 6 |
| chr11_+_66291887 | 0.15 |
ENST00000327259.5
|
TMEM151A
|
transmembrane protein 151A |
| chr11_-_68213277 | 0.15 |
ENST00000401547.6
ENST00000304363.9 ENST00000453170.5 |
KMT5B
|
lysine methyltransferase 5B |
| chr2_+_219434825 | 0.15 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase |
| chr20_-_64079479 | 0.15 |
ENST00000395042.2
|
RGS19
|
regulator of G protein signaling 19 |
| chr6_+_41546340 | 0.15 |
ENST00000307972.10
ENST00000373063.7 |
FOXP4
|
forkhead box P4 |
| chr2_-_177392673 | 0.15 |
ENST00000447413.1
ENST00000397057.6 ENST00000456746.5 ENST00000464747.5 |
ENSG00000213963.6
NFE2L2
|
novel transcript nuclear factor, erythroid 2 like 2 |
| chr5_+_176238365 | 0.15 |
ENST00000341199.10
ENST00000430704.6 ENST00000443967.5 ENST00000429602.7 |
SIMC1
|
SUMO interacting motifs containing 1 |
| chr8_-_73746830 | 0.15 |
ENST00000524300.6
ENST00000523558.5 ENST00000521210.5 ENST00000355780.9 ENST00000524104.5 ENST00000521736.5 ENST00000521447.5 ENST00000517542.5 ENST00000521451.5 ENST00000521419.5 ENST00000518502.5 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr21_+_33543031 | 0.15 |
ENST00000356577.10
ENST00000381692.6 ENST00000300278.8 ENST00000381679.8 |
SON
|
SON DNA and RNA binding protein |
| chr2_+_109794296 | 0.15 |
ENST00000430736.5
ENST00000016946.8 ENST00000441344.1 |
RGPD5
|
RANBP2 like and GRIP domain containing 5 |
| chr16_-_275908 | 0.15 |
ENST00000359740.6
ENST00000316163.9 ENST00000397770.8 |
RGS11
|
regulator of G protein signaling 11 |
| chr8_+_143597814 | 0.15 |
ENST00000504548.4
|
TIGD5
|
tigger transposable element derived 5 |
| chr3_+_191329342 | 0.15 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr22_-_50307598 | 0.15 |
ENST00000425954.1
ENST00000449103.5 ENST00000359337.9 |
PLXNB2
|
plexin B2 |
| chr15_+_92393841 | 0.15 |
ENST00000268164.8
ENST00000539113.5 ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr11_-_27700472 | 0.15 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr5_+_168529299 | 0.15 |
ENST00000338333.5
|
FBLL1
|
fibrillarin like 1 |
| chr8_-_51899002 | 0.14 |
ENST00000522514.6
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr2_-_53786916 | 0.14 |
ENST00000406687.5
ENST00000263634.8 ENST00000394717.3 |
ASB3
|
ankyrin repeat and SOCS box containing 3 |
| chr2_-_110577101 | 0.14 |
ENST00000330331.9
ENST00000446930.1 ENST00000329516.8 |
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr15_-_64775574 | 0.14 |
ENST00000300069.5
|
RBPMS2
|
RNA binding protein, mRNA processing factor 2 |
| chr17_-_44363839 | 0.14 |
ENST00000293443.12
|
FAM171A2
|
family with sequence similarity 171 member A2 |
| chr2_+_45651650 | 0.14 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr19_+_1266653 | 0.14 |
ENST00000586472.5
ENST00000589266.5 |
CIRBP
|
cold inducible RNA binding protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:2000224 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) positive regulation of steroid hormone biosynthetic process(GO:0090031) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 | 2.3 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.2 | 0.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 0.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.6 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.3 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.6 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.6 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.1 | 0.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.4 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.2 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.6 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1901675 | response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0032712 | regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.4 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 1.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.5 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:1903719 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.0 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.1 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 1.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.6 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.2 | 0.6 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 2.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.1 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.4 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.3 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0004935 | adrenergic receptor activity(GO:0004935) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |