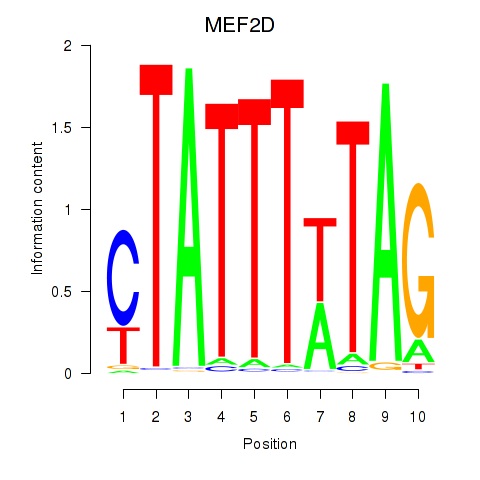
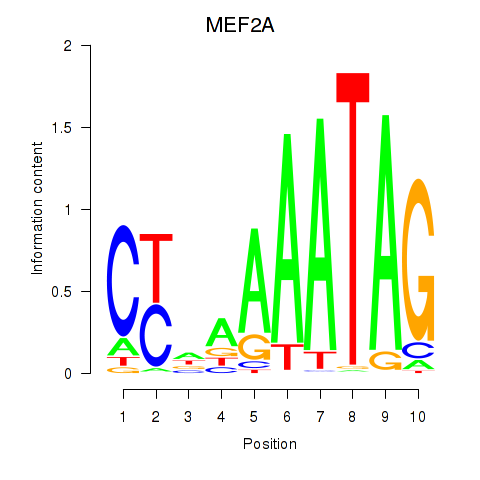
|
chr1_-_152159227
Show fit
|
1.16 |
ENST00000316073.3
|
RPTN
|
repetin
|
|
chr2_-_207165923
Show fit
|
0.59 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7
|
|
chr10_+_116427839
Show fit
|
0.58 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3
|
|
chr11_-_19201976
Show fit
|
0.55 |
ENST00000647990.1
ENST00000649235.1
ENST00000265968.9
ENST00000649842.1
|
CSRP3
|
cysteine and glycine rich protein 3
|
|
chr2_-_207166818
Show fit
|
0.54 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7
|
|
chr11_-_19202004
Show fit
|
0.53 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3
|
|
chr9_+_113594118
Show fit
|
0.51 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3
|
|
chr2_-_189179754
Show fit
|
0.48 |
ENST00000374866.9
ENST00000618828.1
|
COL5A2
|
collagen type V alpha 2 chain
|
|
chr2_-_210303608
Show fit
|
0.47 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1
|
|
chr7_+_143316105
Show fit
|
0.44 |
ENST00000343257.7
ENST00000650516.1
|
CLCN1
|
chloride voltage-gated channel 1
|
|
chr3_+_50279080
Show fit
|
0.43 |
ENST00000316436.4
|
LSMEM2
|
leucine rich single-pass membrane protein 2
|
|
chr11_+_129375841
Show fit
|
0.43 |
ENST00000281437.6
|
BARX2
|
BARX homeobox 2
|
|
chr14_-_64942783
Show fit
|
0.42 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2
|
|
chr14_-_64942720
Show fit
|
0.40 |
ENST00000557049.1
ENST00000389614.6
|
GPX2
|
glutathione peroxidase 2
|
|
chr3_+_138348823
Show fit
|
0.40 |
ENST00000475711.5
ENST00000464896.5
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr5_+_54455661
Show fit
|
0.39 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3
|
|
chr3_+_99638475
Show fit
|
0.38 |
ENST00000452013.5
ENST00000261037.7
ENST00000652472.1
ENST00000273342.8
ENST00000621757.1
|
COL8A1
|
collagen type VIII alpha 1 chain
|
|
chr7_-_113919000
Show fit
|
0.36 |
ENST00000284601.4
|
PPP1R3A
|
protein phosphatase 1 regulatory subunit 3A
|
|
chr3_+_35642159
Show fit
|
0.36 |
ENST00000187397.8
|
ARPP21
|
cAMP regulated phosphoprotein 21
|
|
chr17_+_7281711
Show fit
|
0.36 |
ENST00000317370.13
ENST00000571308.5
|
SLC2A4
|
solute carrier family 2 member 4
|
|
chr2_+_232633551
Show fit
|
0.35 |
ENST00000264059.8
|
EFHD1
|
EF-hand domain family member D1
|
|
chr18_+_12407896
Show fit
|
0.34 |
ENST00000590956.5
ENST00000336990.8
ENST00000440960.6
ENST00000588729.5
|
PRELID3A
|
PRELI domain containing 3A
|
|
chr1_+_150282526
Show fit
|
0.34 |
ENST00000447007.5
ENST00000369095.5
ENST00000369094.5
ENST00000290363.6
|
CIART
|
circadian associated repressor of transcription
|
|
chr7_+_18495723
Show fit
|
0.33 |
ENST00000681950.1
ENST00000622668.4
ENST00000405010.7
ENST00000406451.8
ENST00000441542.7
ENST00000428307.6
ENST00000681273.1
|
HDAC9
|
histone deacetylase 9
|
|
chr1_+_89633086
Show fit
|
0.32 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C
|
|
chr10_-_3785225
Show fit
|
0.32 |
ENST00000542957.1
|
KLF6
|
Kruppel like factor 6
|
|
chr10_-_73655984
Show fit
|
0.31 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like
|
|
chr10_-_3785197
Show fit
|
0.31 |
ENST00000497571.6
|
KLF6
|
Kruppel like factor 6
|
|
chr17_+_44170695
Show fit
|
0.30 |
ENST00000293414.6
|
ASB16
|
ankyrin repeat and SOCS box containing 16
|
|
chr10_-_3785179
Show fit
|
0.30 |
ENST00000469435.1
|
KLF6
|
Kruppel like factor 6
|
|
chr8_+_141417902
Show fit
|
0.29 |
ENST00000681443.1
|
PTP4A3
|
protein tyrosine phosphatase 4A3
|
|
chr2_-_210315160
Show fit
|
0.29 |
ENST00000352451.4
|
MYL1
|
myosin light chain 1
|
|
chr11_+_1838970
Show fit
|
0.29 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr1_-_25430147
Show fit
|
0.29 |
ENST00000349320.7
|
RHCE
|
Rh blood group CcEe antigens
|
|
chr12_+_80099535
Show fit
|
0.28 |
ENST00000646859.1
ENST00000547103.7
|
OTOGL
|
otogelin like
|
|
chr4_-_142305826
Show fit
|
0.27 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr4_-_142846275
Show fit
|
0.27 |
ENST00000513000.5
ENST00000509777.5
ENST00000503927.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr6_-_33192454
Show fit
|
0.26 |
ENST00000395194.1
ENST00000341947.7
ENST00000374708.8
|
COL11A2
|
collagen type XI alpha 2 chain
|
|
chr13_+_108629605
Show fit
|
0.25 |
ENST00000457511.7
|
MYO16
|
myosin XVI
|
|
chr15_-_70702273
Show fit
|
0.25 |
ENST00000558758.5
ENST00000379983.6
ENST00000560441.5
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr8_+_27325516
Show fit
|
0.25 |
ENST00000346049.10
ENST00000420218.3
|
PTK2B
|
protein tyrosine kinase 2 beta
|
|
chr6_+_25726767
Show fit
|
0.25 |
ENST00000274764.5
|
H2BC1
|
H2B clustered histone 1
|
|
chr12_+_4269771
Show fit
|
0.24 |
ENST00000676411.1
|
CCND2
|
cyclin D2
|
|
chr1_+_13585453
Show fit
|
0.24 |
ENST00000487038.5
ENST00000475043.5
|
PDPN
|
podoplanin
|
|
chr7_-_37448845
Show fit
|
0.23 |
ENST00000310758.9
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_+_36210924
Show fit
|
0.23 |
ENST00000615418.4
|
CCL4L2
|
C-C motif chemokine ligand 4 like 2
|
|
chr4_-_142305935
Show fit
|
0.23 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B
|
|
chr17_+_36211055
Show fit
|
0.23 |
ENST00000617405.5
ENST00000617416.4
ENST00000613173.4
ENST00000620732.4
ENST00000620098.4
ENST00000620576.4
ENST00000620055.4
ENST00000610565.4
ENST00000620250.1
|
CCL4L2
|
C-C motif chemokine ligand 4 like 2
|
|
chr5_+_191477
Show fit
|
0.23 |
ENST00000328278.4
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr1_+_84164962
Show fit
|
0.23 |
ENST00000614872.4
ENST00000394839.6
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta
|
|
chr12_-_44875647
Show fit
|
0.22 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2
|
|
chr12_+_15546344
Show fit
|
0.22 |
ENST00000674388.1
ENST00000542557.5
ENST00000445537.6
ENST00000544244.5
ENST00000442921.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O
|
|
chr17_-_74859863
Show fit
|
0.22 |
ENST00000293190.10
|
GRIN2C
|
glutamate ionotropic receptor NMDA type subunit 2C
|
|
chr4_+_119135825
Show fit
|
0.22 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2
|
|
chr2_+_33436304
Show fit
|
0.22 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3
|
|
chr14_+_24132050
Show fit
|
0.22 |
ENST00000559294.1
|
FITM1
|
fat storage inducing transmembrane protein 1
|
|
chr17_-_1486124
Show fit
|
0.22 |
ENST00000575158.5
|
MYO1C
|
myosin IC
|
|
chr19_-_49072699
Show fit
|
0.21 |
ENST00000221444.2
|
KCNA7
|
potassium voltage-gated channel subfamily A member 7
|
|
chr3_+_136022734
Show fit
|
0.21 |
ENST00000334546.6
|
PPP2R3A
|
protein phosphatase 2 regulatory subunit B''alpha
|
|
chr12_-_91146195
Show fit
|
0.21 |
ENST00000548218.1
|
DCN
|
decorin
|
|
chr8_-_7452365
Show fit
|
0.20 |
ENST00000458665.5
ENST00000528168.3
|
SPAG11B
|
sperm associated antigen 11B
|
|
chr9_-_133479075
Show fit
|
0.20 |
ENST00000414172.1
ENST00000371897.8
ENST00000371899.9
|
SLC2A6
|
solute carrier family 2 member 6
|
|
chr2_+_167187283
Show fit
|
0.20 |
ENST00000409605.1
ENST00000409273.6
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr2_-_218831791
Show fit
|
0.19 |
ENST00000439262.6
ENST00000430489.1
|
PRKAG3
|
protein kinase AMP-activated non-catalytic subunit gamma 3
|
|
chr10_-_73651013
Show fit
|
0.19 |
ENST00000372873.8
|
SYNPO2L
|
synaptopodin 2 like
|
|
chrX_-_64230600
Show fit
|
0.19 |
ENST00000362002.3
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr4_+_118888918
Show fit
|
0.18 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2
|
|
chr17_-_1485733
Show fit
|
0.18 |
ENST00000648446.1
|
MYO1C
|
myosin IC
|
|
chr3_+_36380477
Show fit
|
0.18 |
ENST00000457375.6
ENST00000273183.8
ENST00000434649.1
|
STAC
|
SH3 and cysteine rich domain
|
|
chrX_+_79144664
Show fit
|
0.18 |
ENST00000645147.2
|
GPR174
|
G protein-coupled receptor 174
|
|
chr1_+_156126160
Show fit
|
0.18 |
ENST00000448611.6
ENST00000368297.5
|
LMNA
|
lamin A/C
|
|
chr7_-_41703062
Show fit
|
0.18 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A
|
|
chr17_-_29622893
Show fit
|
0.18 |
ENST00000345068.9
ENST00000584602.1
ENST00000388767.8
ENST00000580212.6
|
CORO6
|
coronin 6
|
|
chr1_-_114695613
Show fit
|
0.18 |
ENST00000369538.4
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr9_+_126914844
Show fit
|
0.18 |
ENST00000373436.5
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr7_+_100177743
Show fit
|
0.18 |
ENST00000394018.6
ENST00000416412.5
|
STAG3
|
stromal antigen 3
|
|
chr3_+_138348592
Show fit
|
0.18 |
ENST00000621127.4
ENST00000494949.5
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr2_+_167187364
Show fit
|
0.18 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr22_+_31093358
Show fit
|
0.18 |
ENST00000404574.5
|
SMTN
|
smoothelin
|
|
chr12_-_130716264
Show fit
|
0.18 |
ENST00000643940.1
|
RIMBP2
|
RIMS binding protein 2
|
|
chr17_-_41055211
Show fit
|
0.18 |
ENST00000542910.1
ENST00000398477.1
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr8_+_76683779
Show fit
|
0.18 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr5_-_151141631
Show fit
|
0.18 |
ENST00000523714.5
ENST00000521749.5
|
ANXA6
|
annexin A6
|
|
chr15_-_82806054
Show fit
|
0.17 |
ENST00000541889.1
ENST00000334574.12
ENST00000561368.1
|
FSD2
|
fibronectin type III and SPRY domain containing 2
|
|
chr7_+_100177897
Show fit
|
0.17 |
ENST00000317296.9
ENST00000615138.5
ENST00000620100.5
ENST00000422690.5
ENST00000439782.1
|
STAG3
|
stromal antigen 3
|
|
chr9_+_97501622
Show fit
|
0.17 |
ENST00000259365.9
|
TMOD1
|
tropomodulin 1
|
|
chr10_-_44978789
Show fit
|
0.17 |
ENST00000448778.1
ENST00000298295.4
|
DEPP1
|
DEPP1 autophagy regulator
|
|
chr1_-_114695533
Show fit
|
0.17 |
ENST00000520113.7
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chrX_-_21758097
Show fit
|
0.17 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked
|
|
chr4_+_94451857
Show fit
|
0.17 |
ENST00000318007.9
ENST00000380180.7
ENST00000437932.5
ENST00000615540.4
ENST00000359265.8
ENST00000512274.1
ENST00000503974.5
ENST00000504489.3
ENST00000317968.9
ENST00000542407.5
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr16_+_84294853
Show fit
|
0.17 |
ENST00000219454.10
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr16_+_84294823
Show fit
|
0.16 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr17_-_9791586
Show fit
|
0.16 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C
|
|
chrX_-_15314543
Show fit
|
0.16 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr3_-_108529322
Show fit
|
0.16 |
ENST00000273353.4
|
MYH15
|
myosin heavy chain 15
|
|
chr8_-_27992624
Show fit
|
0.16 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5
|
|
chr5_-_16508812
Show fit
|
0.16 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr5_+_156326735
Show fit
|
0.16 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta
|
|
chr5_-_16508951
Show fit
|
0.16 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr21_-_40847149
Show fit
|
0.16 |
ENST00000400454.6
|
DSCAM
|
DS cell adhesion molecule
|
|
chr11_+_1839602
Show fit
|
0.15 |
ENST00000617947.4
ENST00000252898.11
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr16_+_8674605
Show fit
|
0.15 |
ENST00000268251.13
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_+_1839452
Show fit
|
0.15 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr7_-_44065541
Show fit
|
0.15 |
ENST00000297283.4
|
PGAM2
|
phosphoglycerate mutase 2
|
|
chr20_+_46029206
Show fit
|
0.15 |
ENST00000243964.7
|
SLC12A5
|
solute carrier family 12 member 5
|
|
chr17_-_42979993
Show fit
|
0.15 |
ENST00000409103.5
ENST00000360221.8
ENST00000454303.1
ENST00000591916.7
ENST00000451885.3
|
PTGES3L-AARSD1
PTGES3L
|
PTGES3L-AARSD1 readthrough
prostaglandin E synthase 3 like
|
|
chr7_+_18496269
Show fit
|
0.15 |
ENST00000432645.6
|
HDAC9
|
histone deacetylase 9
|
|
chr5_-_16508858
Show fit
|
0.15 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr1_+_26021768
Show fit
|
0.15 |
ENST00000374280.4
|
EXTL1
|
exostosin like glycosyltransferase 1
|
|
chr20_+_46029165
Show fit
|
0.14 |
ENST00000616201.4
ENST00000616202.4
ENST00000616933.4
ENST00000626937.2
|
SLC12A5
|
solute carrier family 12 member 5
|
|
chr12_+_119178920
Show fit
|
0.14 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8
|
|
chr8_+_103819244
Show fit
|
0.14 |
ENST00000262231.14
ENST00000507740.5
ENST00000408894.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr6_+_131808011
Show fit
|
0.14 |
ENST00000647893.1
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr17_-_42980393
Show fit
|
0.14 |
ENST00000409446.8
ENST00000409399.6
ENST00000421990.7
|
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 like
PTGES3L-AARSD1 readthrough
|
|
chr3_-_30894661
Show fit
|
0.13 |
ENST00000282538.10
ENST00000454381.3
|
GADL1
|
glutamate decarboxylase like 1
|
|
chr3_-_47892743
Show fit
|
0.13 |
ENST00000420772.6
|
MAP4
|
microtubule associated protein 4
|
|
chr7_+_154305105
Show fit
|
0.13 |
ENST00000332007.7
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr2_+_88067818
Show fit
|
0.13 |
ENST00000444564.2
ENST00000419482.7
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr5_-_95682968
Show fit
|
0.13 |
ENST00000274432.13
|
SPATA9
|
spermatogenesis associated 9
|
|
chr19_-_47471847
Show fit
|
0.13 |
ENST00000594353.1
ENST00000542837.2
|
SLC8A2
|
solute carrier family 8 member A2
|
|
chr12_+_40224956
Show fit
|
0.13 |
ENST00000298910.12
ENST00000343742.6
|
LRRK2
|
leucine rich repeat kinase 2
|
|
chr19_-_46661132
Show fit
|
0.13 |
ENST00000410105.2
ENST00000391916.7
|
DACT3
|
dishevelled binding antagonist of beta catenin 3
|
|
chr12_+_119178953
Show fit
|
0.13 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8
|
|
chr5_-_132011811
Show fit
|
0.13 |
ENST00000379255.5
ENST00000430403.5
ENST00000357096.5
|
ACSL6
|
acyl-CoA synthetase long chain family member 6
|
|
chr8_-_27258414
Show fit
|
0.13 |
ENST00000523048.5
|
STMN4
|
stathmin 4
|
|
chr14_-_56810448
Show fit
|
0.13 |
ENST00000339475.10
ENST00000555006.5
ENST00000672264.2
ENST00000554559.5
ENST00000555804.1
|
OTX2
|
orthodenticle homeobox 2
|
|
chr6_-_27838362
Show fit
|
0.13 |
ENST00000618958.2
|
H2AC15
|
H2A clustered histone 15
|
|
chr19_-_47471886
Show fit
|
0.13 |
ENST00000236877.11
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 member A2
|
|
chr17_+_39665340
Show fit
|
0.13 |
ENST00000578283.1
ENST00000309889.3
|
TCAP
|
titin-cap
|
|
chr17_-_36017953
Show fit
|
0.12 |
ENST00000612516.4
ENST00000615050.2
|
CCL23
|
C-C motif chemokine ligand 23
|
|
chr3_-_57292676
Show fit
|
0.12 |
ENST00000389601.3
ENST00000487349.6
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr19_+_45469841
Show fit
|
0.12 |
ENST00000592811.5
ENST00000586615.5
|
FOSB
|
FosB proto-oncogene, AP-1 transcription factor subunit
|
|
chr12_+_53954870
Show fit
|
0.12 |
ENST00000243103.4
|
HOXC12
|
homeobox C12
|
|
chr3_+_138348570
Show fit
|
0.12 |
ENST00000423968.7
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr1_-_100178215
Show fit
|
0.12 |
ENST00000370138.1
ENST00000370137.6
ENST00000342895.7
ENST00000620882.4
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr1_-_1000139
Show fit
|
0.12 |
ENST00000428771.6
|
HES4
|
hes family bHLH transcription factor 4
|
|
chr2_+_113059194
Show fit
|
0.12 |
ENST00000393200.7
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr21_-_32813695
Show fit
|
0.12 |
ENST00000479548.2
ENST00000490358.5
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr8_-_42377227
Show fit
|
0.12 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4
|
|
chr11_+_112961402
Show fit
|
0.12 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_-_262828
Show fit
|
0.12 |
ENST00000343164.9
ENST00000436453.1
ENST00000445055.6
ENST00000546319.5
|
SLC6A13
|
solute carrier family 6 member 13
|
|
chr11_-_31803960
Show fit
|
0.11 |
ENST00000640872.1
ENST00000639109.1
ENST00000638629.1
ENST00000639386.2
|
PAX6
|
paired box 6
|
|
chr20_-_45891200
Show fit
|
0.11 |
ENST00000372518.5
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2
|
|
chr14_-_69574845
Show fit
|
0.11 |
ENST00000599174.3
|
CCDC177
|
coiled-coil domain containing 177
|
|
chr1_+_175067831
Show fit
|
0.11 |
ENST00000239462.9
|
TNN
|
tenascin N
|
|
chr11_+_112961480
Show fit
|
0.11 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr15_-_52528821
Show fit
|
0.11 |
ENST00000553916.5
|
MYO5A
|
myosin VA
|
|
chr9_+_5450503
Show fit
|
0.11 |
ENST00000381573.8
ENST00000381577.4
|
CD274
|
CD274 molecule
|
|
chr17_-_10421944
Show fit
|
0.11 |
ENST00000403437.2
|
MYH8
|
myosin heavy chain 8
|
|
chr11_+_3645105
Show fit
|
0.11 |
ENST00000250693.2
|
ART1
|
ADP-ribosyltransferase 1
|
|
chr9_+_136977496
Show fit
|
0.11 |
ENST00000371625.8
ENST00000457950.5
|
PTGDS
|
prostaglandin D2 synthase
|
|
chr6_-_123636979
Show fit
|
0.11 |
ENST00000662930.1
|
TRDN
|
triadin
|
|
chr4_+_118888829
Show fit
|
0.11 |
ENST00000448416.6
ENST00000307142.9
ENST00000429713.7
|
SYNPO2
|
synaptopodin 2
|
|
chr4_-_121381007
Show fit
|
0.11 |
ENST00000394427.3
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr2_+_104853259
Show fit
|
0.10 |
ENST00000598623.1
ENST00000653688.1
ENST00000662784.1
ENST00000666977.1
ENST00000674056.1
|
ENSG00000269707.2
POU3F3
|
novel transcript, sense overlapping POU3F3
POU class 3 homeobox 3
|
|
chr19_+_6739650
Show fit
|
0.10 |
ENST00000313285.12
ENST00000313244.14
ENST00000596758.5
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr1_+_113979391
Show fit
|
0.10 |
ENST00000393300.6
ENST00000369551.5
|
OLFML3
|
olfactomedin like 3
|
|
chr2_+_112645930
Show fit
|
0.10 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1
|
|
chr3_+_66998297
Show fit
|
0.10 |
ENST00000484414.5
ENST00000460576.5
ENST00000417314.2
|
KBTBD8
|
kelch repeat and BTB domain containing 8
|
|
chr17_+_7439504
Show fit
|
0.10 |
ENST00000575331.1
ENST00000293829.9
|
ENSG00000272884.1
FGF11
|
novel transcript
fibroblast growth factor 11
|
|
chr6_+_44227025
Show fit
|
0.10 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group)
|
|
chr1_+_113979460
Show fit
|
0.10 |
ENST00000320334.5
|
OLFML3
|
olfactomedin like 3
|
|
chr11_-_31804067
Show fit
|
0.10 |
ENST00000639548.1
ENST00000640125.1
ENST00000481563.6
ENST00000639079.1
ENST00000638762.1
ENST00000638346.1
|
PAX6
|
paired box 6
|
|
chr18_+_23135452
Show fit
|
0.10 |
ENST00000580153.5
ENST00000256925.12
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr17_+_17782108
Show fit
|
0.10 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1
|
|
chr17_+_7855055
Show fit
|
0.10 |
ENST00000574668.1
ENST00000301599.7
|
TMEM88
|
transmembrane protein 88
|
|
chr2_+_44275457
Show fit
|
0.10 |
ENST00000611973.4
ENST00000409387.5
|
SLC3A1
|
solute carrier family 3 member 1
|
|
chr10_-_67665642
Show fit
|
0.09 |
ENST00000682945.1
ENST00000330298.6
ENST00000682758.1
|
CTNNA3
|
catenin alpha 3
|
|
chr6_+_151807319
Show fit
|
0.09 |
ENST00000443427.5
|
ESR1
|
estrogen receptor 1
|
|
chr7_-_78771108
Show fit
|
0.09 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_-_35420017
Show fit
|
0.09 |
ENST00000643000.1
ENST00000646099.1
ENST00000647372.1
ENST00000642578.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr1_+_160081529
Show fit
|
0.09 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9
|
|
chr2_+_219434825
Show fit
|
0.09 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase
|
|
chr14_-_103522696
Show fit
|
0.09 |
ENST00000553878.5
ENST00000348956.7
ENST00000557530.1
|
CKB
|
creatine kinase B
|
|
chr16_+_88382950
Show fit
|
0.09 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469
|
|
chr1_-_205943449
Show fit
|
0.09 |
ENST00000367135.8
ENST00000367134.2
|
SLC26A9
|
solute carrier family 26 member 9
|
|
chr11_+_124611420
Show fit
|
0.09 |
ENST00000284288.3
|
PANX3
|
pannexin 3
|
|
chr9_-_127877665
Show fit
|
0.09 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1
|
|
chr18_+_44680093
Show fit
|
0.09 |
ENST00000426838.8
ENST00000677068.1
|
SETBP1
|
SET binding protein 1
|
|
chr4_+_37891060
Show fit
|
0.09 |
ENST00000261439.9
ENST00000508802.5
ENST00000402522.1
|
TBC1D1
|
TBC1 domain family member 1
|
|
chr20_+_45407207
Show fit
|
0.08 |
ENST00000372712.6
|
DBNDD2
|
dysbindin domain containing 2
|
|
chr11_-_35419213
Show fit
|
0.08 |
ENST00000642171.1
ENST00000644050.1
ENST00000643134.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr11_-_64759967
Show fit
|
0.08 |
ENST00000377432.7
ENST00000164139.4
|
PYGM
|
glycogen phosphorylase, muscle associated
|
|
chr3_+_133124807
Show fit
|
0.08 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108
|
|
chr7_+_95485934
Show fit
|
0.08 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr10_+_84424919
Show fit
|
0.08 |
ENST00000543283.2
ENST00000494586.5
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr5_-_142698004
Show fit
|
0.08 |
ENST00000407758.5
ENST00000441680.6
ENST00000419524.6
ENST00000621536.4
|
FGF1
|
fibroblast growth factor 1
|
|
chr18_-_37565825
Show fit
|
0.08 |
ENST00000603232.6
|
CELF4
|
CUGBP Elav-like family member 4
|
|
chr16_+_11965193
Show fit
|
0.08 |
ENST00000053243.6
ENST00000396495.3
|
TNFRSF17
|
TNF receptor superfamily member 17
|
|
chr1_+_150272772
Show fit
|
0.08 |
ENST00000369098.3
ENST00000369099.8
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr7_+_154305256
Show fit
|
0.08 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr8_+_73991345
Show fit
|
0.08 |
ENST00000284818.7
ENST00000518893.1
|
LY96
|
lymphocyte antigen 96
|
|
chr12_+_101594849
Show fit
|
0.08 |
ENST00000547405.5
ENST00000452455.6
ENST00000392934.7
ENST00000547509.5
ENST00000361685.6
ENST00000549145.5
ENST00000361466.7
ENST00000553190.5
ENST00000545503.6
ENST00000536007.5
ENST00000541119.5
ENST00000551300.5
ENST00000550270.1
|
MYBPC1
|
myosin binding protein C1
|
|
chr4_-_139177185
Show fit
|
0.08 |
ENST00000394235.6
|
ELF2
|
E74 like ETS transcription factor 2
|
|
chr6_+_54083423
Show fit
|
0.08 |
ENST00000460844.6
ENST00000370876.6
|
MLIP
|
muscular LMNA interacting protein
|
|
chr19_+_53991630
Show fit
|
0.08 |
ENST00000252729.7
|
CACNG6
|
calcium voltage-gated channel auxiliary subunit gamma 6
|
|
chr1_-_169734064
Show fit
|
0.08 |
ENST00000333360.12
|
SELE
|
selectin E
|
|
chr11_-_35419098
Show fit
|
0.08 |
ENST00000606205.6
ENST00000645303.1
|
SLC1A2
|
solute carrier family 1 member 2
|
|
chr13_-_102759059
Show fit
|
0.08 |
ENST00000322527.4
|
CCDC168
|
coiled-coil domain containing 168
|
|
chr2_+_238138661
Show fit
|
0.08 |
ENST00000409223.2
|
KLHL30
|
kelch like family member 30
|