Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
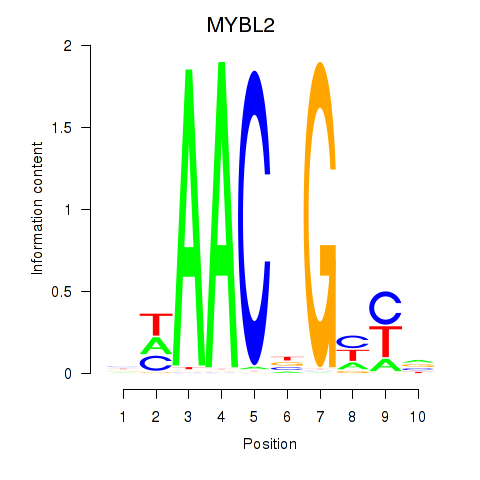
Results for MYBL2
Z-value: 1.88
Transcription factors associated with MYBL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL2
|
ENSG00000101057.16 | MYBL2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL2 | hg38_v1_chr20_+_43667105_43667125, hg38_v1_chr20_+_43667019_43667098 | 0.01 | 9.7e-01 | Click! |
Activity profile of MYBL2 motif
Sorted Z-values of MYBL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 1.3 | 5.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 1.0 | 3.0 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.9 | 18.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.8 | 8.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.8 | 2.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.8 | 13.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.8 | 2.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.7 | 5.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.7 | 2.1 | GO:1904868 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.7 | 3.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.7 | 3.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.6 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 12.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.5 | 1.6 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.5 | 3.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.4 | 8.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.4 | 0.4 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.4 | 6.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.4 | 1.6 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.3 | 1.7 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 0.9 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 0.9 | GO:0006212 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.3 | 0.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 0.9 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.3 | 2.3 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.3 | 0.8 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.3 | 3.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 0.8 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.3 | 2.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 0.5 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.2 | 0.7 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.2 | 2.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.8 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.8 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 1.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.2 | 0.4 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.2 | 1.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 3.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.9 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 1.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 0.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 1.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 1.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.7 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.4 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 0.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.1 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 5.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.5 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.9 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 3.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 3.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.4 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 2.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.6 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 1.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 3.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 2.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 1.5 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.7 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.7 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 2.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.5 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) regulation of metanephric glomerular mesangial cell proliferation(GO:0072301) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.3 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.3 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 6.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 6.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.6 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 2.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.4 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 1.1 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 1.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:0009196 | dUDP biosynthetic process(GO:0006227) dTDP biosynthetic process(GO:0006233) dTTP biosynthetic process(GO:0006235) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTDP metabolic process(GO:0046072) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.0 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 1.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.1 | GO:0090042 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.5 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.8 | 2.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.7 | 2.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.7 | 8.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.7 | 7.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.7 | 4.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 3.8 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 2.0 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.5 | 2.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.4 | 1.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.4 | 16.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 7.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.5 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 4.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 2.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 2.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 0.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.2 | 0.8 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 10.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 3.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 7.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 3.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 2.5 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.4 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 12.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 2.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 2.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 4.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 2.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 2.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 13.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 3.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.6 | 2.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.6 | 2.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.6 | 4.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 6.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.5 | 2.3 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.5 | 2.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.4 | 3.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 1.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.4 | 1.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 0.6 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.3 | 1.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 1.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 0.8 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.2 | 1.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 2.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 0.7 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.2 | 2.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 5.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 0.8 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.7 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 4.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 0.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 2.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0004618 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 1.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 3.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 4.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.6 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.5 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 2.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 3.8 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 2.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.3 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 3.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 6.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.7 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 3.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 5.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 3.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 10.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 4.0 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 3.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |