Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
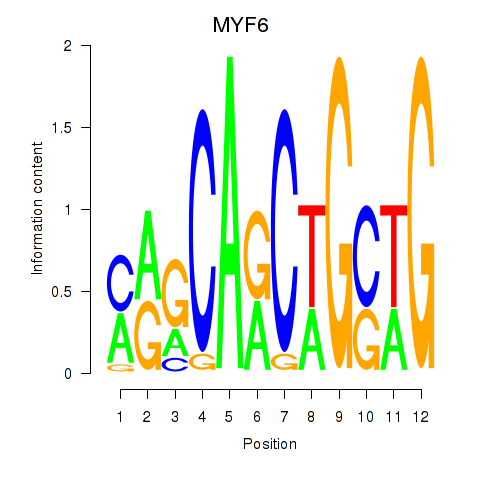
Results for MYF6
Z-value: 1.02
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.4 | MYF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg38_v1_chr12_+_80707625_80707642 | 0.30 | 1.1e-01 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_41118369 | 6.91 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr12_-_76031588 | 5.01 |
ENST00000602540.5
|
PHLDA1
|
pleckstrin homology like domain family A member 1 |
| chr19_-_51001138 | 4.32 |
ENST00000593490.1
|
KLK8
|
kallikrein related peptidase 8 |
| chrX_-_38220824 | 3.54 |
ENST00000378533.4
ENST00000432886.6 ENST00000544439.5 ENST00000538295.5 |
SRPX
|
sushi repeat containing protein X-linked |
| chr1_-_153070840 | 3.17 |
ENST00000368755.2
|
SPRR2B
|
small proline rich protein 2B |
| chr19_-_51001591 | 3.16 |
ENST00000391806.6
|
KLK8
|
kallikrein related peptidase 8 |
| chr20_-_54173976 | 2.79 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr20_-_54173516 | 2.77 |
ENST00000395955.7
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr1_+_20589044 | 2.29 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr17_-_41098084 | 2.26 |
ENST00000318329.6
ENST00000333822.5 |
KRTAP4-8
|
keratin associated protein 4-8 |
| chr2_-_215436061 | 2.00 |
ENST00000421182.5
ENST00000432072.6 ENST00000323926.10 ENST00000336916.8 ENST00000357867.8 ENST00000359671.5 ENST00000446046.5 ENST00000354785.11 ENST00000356005.8 ENST00000443816.5 ENST00000426059.1 |
FN1
|
fibronectin 1 |
| chr19_+_53867874 | 1.80 |
ENST00000448420.5
ENST00000439000.5 ENST00000391771.1 ENST00000391770.9 |
MYADM
|
myeloid associated differentiation marker |
| chr22_+_31093358 | 1.71 |
ENST00000404574.5
|
SMTN
|
smoothelin |
| chr17_-_41168219 | 1.70 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr20_+_35172046 | 1.68 |
ENST00000216968.5
|
PROCR
|
protein C receptor |
| chr14_+_32934383 | 1.65 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr4_+_7192519 | 1.65 |
ENST00000507866.6
|
SORCS2
|
sortilin related VPS10 domain containing receptor 2 |
| chr9_+_122371036 | 1.63 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr20_+_3786772 | 1.61 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr9_+_113536497 | 1.60 |
ENST00000462143.5
|
RGS3
|
regulator of G protein signaling 3 |
| chr17_-_41149823 | 1.56 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr14_-_74955577 | 1.51 |
ENST00000238607.10
ENST00000555567.6 ENST00000553716.5 |
PGF
|
placental growth factor |
| chr13_-_35476682 | 1.38 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1 |
| chr19_+_917287 | 1.37 |
ENST00000592648.1
ENST00000234371.10 |
KISS1R
|
KISS1 receptor |
| chr17_-_41350824 | 1.34 |
ENST00000007735.4
|
KRT33A
|
keratin 33A |
| chr6_+_37170133 | 1.33 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr9_-_35685462 | 1.32 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 |
| chr6_-_30687200 | 1.27 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr17_-_41382298 | 1.27 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr19_-_18940289 | 1.25 |
ENST00000542541.6
ENST00000433218.6 |
HOMER3
|
homer scaffold protein 3 |
| chr19_-_37906646 | 1.22 |
ENST00000303868.5
|
WDR87
|
WD repeat domain 87 |
| chr1_-_16978276 | 1.20 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr11_-_65557740 | 1.17 |
ENST00000526927.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr8_+_22165140 | 1.16 |
ENST00000397814.7
ENST00000354870.5 |
BMP1
|
bone morphogenetic protein 1 |
| chr19_-_15232399 | 1.16 |
ENST00000221730.8
|
EPHX3
|
epoxide hydrolase 3 |
| chr9_+_122371014 | 1.15 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr8_+_31639755 | 1.15 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr13_+_110307276 | 1.13 |
ENST00000360467.7
ENST00000650540.1 |
COL4A2
|
collagen type IV alpha 2 chain |
| chr17_+_41812974 | 1.12 |
ENST00000321562.9
|
FKBP10
|
FKBP prolyl isomerase 10 |
| chr8_+_22165358 | 1.12 |
ENST00000306349.13
ENST00000306385.10 |
BMP1
|
bone morphogenetic protein 1 |
| chrX_-_46759055 | 1.11 |
ENST00000328306.4
ENST00000616978.5 |
SLC9A7
|
solute carrier family 9 member A7 |
| chr16_-_2720217 | 1.11 |
ENST00000302641.8
|
PRSS27
|
serine protease 27 |
| chr11_-_1622093 | 1.11 |
ENST00000616115.1
ENST00000399682.1 |
KRTAP5-4
|
keratin associated protein 5-4 |
| chr1_+_152985231 | 1.10 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr10_-_77637902 | 1.09 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr9_+_130053897 | 1.08 |
ENST00000347136.11
ENST00000610997.1 |
GPR107
|
G protein-coupled receptor 107 |
| chr9_+_130053706 | 1.05 |
ENST00000372410.7
|
GPR107
|
G protein-coupled receptor 107 |
| chr21_+_36135071 | 1.05 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3 |
| chr20_-_62367304 | 1.04 |
ENST00000252999.7
|
LAMA5
|
laminin subunit alpha 5 |
| chr1_-_9129085 | 1.04 |
ENST00000377411.5
|
GPR157
|
G protein-coupled receptor 157 |
| chr19_-_50511203 | 1.02 |
ENST00000595669.5
|
JOSD2
|
Josephin domain containing 2 |
| chr21_+_29130630 | 1.02 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr17_-_41369807 | 1.01 |
ENST00000251646.8
|
KRT33B
|
keratin 33B |
| chr17_+_41255384 | 1.01 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr9_+_35673917 | 1.01 |
ENST00000617161.1
ENST00000378357.9 |
CA9
|
carbonic anhydrase 9 |
| chr5_-_147782518 | 1.01 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr1_+_55039511 | 0.97 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr11_-_66958366 | 0.97 |
ENST00000651036.1
ENST00000652125.1 ENST00000531614.6 ENST00000524491.6 ENST00000529047.6 ENST00000393960.7 ENST00000393958.7 ENST00000528403.6 ENST00000651854.1 |
PC
|
pyruvate carboxylase |
| chr5_-_140633167 | 0.96 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr18_+_58863580 | 0.96 |
ENST00000586085.5
ENST00000589288.5 |
ZNF532
|
zinc finger protein 532 |
| chrX_-_107775740 | 0.96 |
ENST00000372383.9
|
TSC22D3
|
TSC22 domain family member 3 |
| chr16_+_30053123 | 0.95 |
ENST00000395248.6
ENST00000575627.5 ENST00000566897.6 ENST00000568435.6 ENST00000338110.11 ENST00000562240.1 |
ENSG00000285043.3
|
novel protein |
| chr2_+_148021083 | 0.95 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr17_-_41397600 | 0.95 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr13_-_113864062 | 0.94 |
ENST00000327773.7
|
GAS6
|
growth arrest specific 6 |
| chr17_+_76385256 | 0.94 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chrX_-_107716401 | 0.93 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr15_-_63381835 | 0.93 |
ENST00000344366.7
ENST00000178638.8 ENST00000422263.2 |
CA12
|
carbonic anhydrase 12 |
| chr11_+_71538025 | 0.92 |
ENST00000398534.4
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr19_+_39196956 | 0.92 |
ENST00000339852.5
|
NCCRP1
|
NCCRP1, F-box associated domain containing |
| chr18_-_23586422 | 0.91 |
ENST00000269228.10
|
NPC1
|
NPC intracellular cholesterol transporter 1 |
| chr2_+_148021404 | 0.91 |
ENST00000638043.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr11_+_134069060 | 0.91 |
ENST00000534549.5
ENST00000441717.3 ENST00000299106.9 |
JAM3
|
junctional adhesion molecule 3 |
| chr1_+_32222393 | 0.90 |
ENST00000676679.1
ENST00000678689.1 ENST00000373586.2 ENST00000679290.1 ENST00000677378.1 ENST00000678534.1 ENST00000678968.1 ENST00000678063.1 ENST00000678150.1 ENST00000677198.1 ENST00000677540.1 |
EIF3I
|
eukaryotic translation initiation factor 3 subunit I |
| chr11_-_65558338 | 0.90 |
ENST00000301873.11
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr5_-_147782681 | 0.89 |
ENST00000616793.5
ENST00000333010.6 ENST00000265272.9 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr19_+_6531018 | 0.89 |
ENST00000245817.5
|
TNFSF9
|
TNF superfamily member 9 |
| chr2_+_148021001 | 0.89 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr2_+_114442616 | 0.89 |
ENST00000410059.6
|
DPP10
|
dipeptidyl peptidase like 10 |
| chr2_+_6865557 | 0.88 |
ENST00000680607.1
ENST00000680320.1 ENST00000442639.6 |
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr17_+_2056073 | 0.88 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr11_-_62556230 | 0.88 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr12_-_54419259 | 0.87 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr13_+_108629605 | 0.87 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr7_+_2647703 | 0.85 |
ENST00000403167.5
|
TTYH3
|
tweety family member 3 |
| chr3_-_125055987 | 0.84 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr8_-_67746348 | 0.83 |
ENST00000297770.10
|
CPA6
|
carboxypeptidase A6 |
| chr19_+_54415427 | 0.82 |
ENST00000301194.8
ENST00000376530.8 ENST00000445095.5 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr17_-_7393404 | 0.81 |
ENST00000575434.4
|
PLSCR3
|
phospholipid scramblase 3 |
| chr10_-_77638369 | 0.79 |
ENST00000372443.6
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr2_+_190880834 | 0.79 |
ENST00000338435.8
|
GLS
|
glutaminase |
| chr4_-_55636259 | 0.79 |
ENST00000505262.5
ENST00000507338.1 |
NMU
|
neuromedin U |
| chr3_-_58211212 | 0.79 |
ENST00000461914.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr10_+_97713694 | 0.78 |
ENST00000285605.8
|
MARVELD1
|
MARVEL domain containing 1 |
| chr8_-_28386073 | 0.78 |
ENST00000523095.5
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr19_+_38319807 | 0.78 |
ENST00000263372.5
|
KCNK6
|
potassium two pore domain channel subfamily K member 6 |
| chrX_-_120560884 | 0.78 |
ENST00000404115.8
|
CUL4B
|
cullin 4B |
| chr9_+_136662907 | 0.77 |
ENST00000308874.12
ENST00000406555.7 ENST00000492862.6 |
EGFL7
|
EGF like domain multiple 7 |
| chr5_+_93584916 | 0.77 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr2_+_37344594 | 0.76 |
ENST00000404976.5
ENST00000338415.8 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr3_-_86991135 | 0.74 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3 |
| chr5_-_36241798 | 0.73 |
ENST00000514504.5
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr17_+_8002610 | 0.73 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal |
| chr2_+_190880809 | 0.73 |
ENST00000320717.8
|
GLS
|
glutaminase |
| chr19_+_46601237 | 0.72 |
ENST00000597743.5
|
CALM3
|
calmodulin 3 |
| chr11_-_14891643 | 0.72 |
ENST00000532378.5
|
CYP2R1
|
cytochrome P450 family 2 subfamily R member 1 |
| chr6_+_7726089 | 0.71 |
ENST00000283147.7
|
BMP6
|
bone morphogenetic protein 6 |
| chr3_-_52826834 | 0.70 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr8_+_32548210 | 0.70 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr19_-_45405034 | 0.69 |
ENST00000592134.1
ENST00000360957.10 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr14_+_73239599 | 0.68 |
ENST00000554301.5
ENST00000555445.5 |
PAPLN
|
papilin, proteoglycan like sulfated glycoprotein |
| chr16_+_2817230 | 0.67 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr15_+_90201301 | 0.67 |
ENST00000411539.6
|
SEMA4B
|
semaphorin 4B |
| chr16_+_2817180 | 0.67 |
ENST00000450020.7
|
PRSS21
|
serine protease 21 |
| chr1_+_155613221 | 0.67 |
ENST00000462250.2
|
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr4_-_55636284 | 0.66 |
ENST00000511469.5
ENST00000264218.7 |
NMU
|
neuromedin U |
| chr10_-_89207306 | 0.66 |
ENST00000371852.4
|
CH25H
|
cholesterol 25-hydroxylase |
| chrX_+_56232411 | 0.66 |
ENST00000374928.7
|
KLF8
|
Kruppel like factor 8 |
| chrX_-_111270474 | 0.66 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr11_-_14892217 | 0.66 |
ENST00000334636.10
|
CYP2R1
|
cytochrome P450 family 2 subfamily R member 1 |
| chr8_-_28386417 | 0.65 |
ENST00000521185.5
ENST00000520290.5 ENST00000344423.10 |
ZNF395
|
zinc finger protein 395 |
| chr12_-_32896757 | 0.65 |
ENST00000070846.11
ENST00000340811.9 |
PKP2
|
plakophilin 2 |
| chr9_-_98708856 | 0.65 |
ENST00000259455.4
|
GABBR2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr7_-_101217569 | 0.64 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr12_+_57128475 | 0.64 |
ENST00000243077.8
ENST00000553277.5 |
LRP1
|
LDL receptor related protein 1 |
| chr15_-_74433942 | 0.64 |
ENST00000543145.6
ENST00000261918.9 |
SEMA7A
|
semaphorin 7A (John Milton Hagen blood group) |
| chr17_-_41027198 | 0.63 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr17_-_41055211 | 0.63 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr20_+_38926312 | 0.63 |
ENST00000619304.4
ENST00000619850.2 |
FAM83D
|
family with sequence similarity 83 member D |
| chr2_+_238100332 | 0.63 |
ENST00000343063.8
|
ESPNL
|
espin like |
| chr22_+_50705495 | 0.62 |
ENST00000664402.1
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr17_-_41811930 | 0.62 |
ENST00000393928.6
|
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr1_+_32222415 | 0.62 |
ENST00000678420.1
ENST00000678162.1 ENST00000678711.1 ENST00000678883.1 ENST00000677353.1 ENST00000355082.10 |
EIF3I
|
eukaryotic translation initiation factor 3 subunit I |
| chr17_-_41612757 | 0.62 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr14_+_24398986 | 0.61 |
ENST00000382554.4
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr19_-_43670153 | 0.60 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_+_117179218 | 0.60 |
ENST00000628876.2
ENST00000431081.6 |
SIDT2
|
SID1 transmembrane family member 2 |
| chr4_-_102345061 | 0.60 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8 |
| chr21_+_37367097 | 0.60 |
ENST00000644942.1
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr2_+_238100393 | 0.60 |
ENST00000409169.5
|
ESPNL
|
espin like |
| chr6_-_127459364 | 0.60 |
ENST00000487331.2
ENST00000483725.8 |
KIAA0408
|
KIAA0408 |
| chr6_-_32154326 | 0.59 |
ENST00000475826.1
ENST00000485392.5 ENST00000494332.5 ENST00000498575.1 ENST00000428778.5 |
ENSG00000284954.1
ENSG00000285085.1
|
novel transcript novel protein |
| chr3_-_73624840 | 0.59 |
ENST00000308537.4
ENST00000263666.9 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr5_-_73448769 | 0.59 |
ENST00000615637.3
|
FOXD1
|
forkhead box D1 |
| chr4_-_56977577 | 0.59 |
ENST00000264230.5
|
NOA1
|
nitric oxide associated 1 |
| chr1_-_230978796 | 0.59 |
ENST00000522821.5
ENST00000366662.8 ENST00000366661.9 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr11_+_117179127 | 0.59 |
ENST00000278951.11
|
SIDT2
|
SID1 transmembrane family member 2 |
| chr2_-_25982471 | 0.58 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr9_+_4985227 | 0.58 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2 |
| chr19_+_53882186 | 0.58 |
ENST00000682028.1
ENST00000683513.1 ENST00000263431.4 ENST00000419486.1 |
PRKCG
|
protein kinase C gamma |
| chr12_-_95116967 | 0.58 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr17_+_50056095 | 0.57 |
ENST00000320031.13
|
ITGA3
|
integrin subunit alpha 3 |
| chr19_+_639895 | 0.57 |
ENST00000586042.6
ENST00000215530.6 |
FGF22
|
fibroblast growth factor 22 |
| chr4_-_41214450 | 0.57 |
ENST00000513140.5
|
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr14_-_64823148 | 0.57 |
ENST00000389722.7
|
SPTB
|
spectrin beta, erythrocytic |
| chr6_+_45422485 | 0.56 |
ENST00000359524.7
|
RUNX2
|
RUNX family transcription factor 2 |
| chr4_-_52038260 | 0.56 |
ENST00000381431.10
|
SGCB
|
sarcoglycan beta |
| chr19_-_46023046 | 0.55 |
ENST00000008938.5
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr11_-_67353503 | 0.55 |
ENST00000539074.1
ENST00000530584.5 ENST00000531239.2 ENST00000312419.8 ENST00000529704.5 |
POLD4
|
DNA polymerase delta 4, accessory subunit |
| chr2_+_10122730 | 0.55 |
ENST00000304567.10
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr3_-_190120881 | 0.55 |
ENST00000319332.10
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr22_+_18527802 | 0.54 |
ENST00000612978.5
|
TMEM191B
|
transmembrane protein 191B |
| chr17_-_82051800 | 0.54 |
ENST00000310496.9
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_+_131289685 | 0.54 |
ENST00000372264.4
|
PLPP7
|
phospholipid phosphatase 7 (inactive) |
| chr11_+_65890627 | 0.54 |
ENST00000312579.4
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr20_+_2814981 | 0.54 |
ENST00000603872.2
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr17_-_41586887 | 0.53 |
ENST00000167586.7
|
KRT14
|
keratin 14 |
| chr16_+_2019777 | 0.53 |
ENST00000566435.4
|
NPW
|
neuropeptide W |
| chr10_-_77637558 | 0.52 |
ENST00000372421.10
ENST00000639370.1 ENST00000640773.1 ENST00000638895.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr11_-_627173 | 0.52 |
ENST00000176195.4
|
SCT
|
secretin |
| chr12_+_56128217 | 0.52 |
ENST00000267113.4
ENST00000394048.10 |
ESYT1
|
extended synaptotagmin 1 |
| chr12_-_84912816 | 0.52 |
ENST00000680469.1
ENST00000450363.4 ENST00000681106.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_-_197260369 | 0.52 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr5_+_138179093 | 0.52 |
ENST00000394894.8
|
KIF20A
|
kinesin family member 20A |
| chrX_-_132961390 | 0.52 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr7_+_31687208 | 0.51 |
ENST00000409146.3
ENST00000342032.8 |
PPP1R17
|
protein phosphatase 1 regulatory subunit 17 |
| chrX_+_56232343 | 0.51 |
ENST00000468660.6
|
KLF8
|
Kruppel like factor 8 |
| chr8_-_4994696 | 0.51 |
ENST00000400186.7
ENST00000602723.5 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr9_-_112175283 | 0.51 |
ENST00000374270.8
ENST00000374263.7 |
SUSD1
|
sushi domain containing 1 |
| chr1_-_6491664 | 0.51 |
ENST00000377728.8
ENST00000675093.1 |
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chrX_+_87517784 | 0.51 |
ENST00000373119.9
ENST00000373114.4 |
KLHL4
|
kelch like family member 4 |
| chr2_-_74440484 | 0.51 |
ENST00000305557.9
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr12_+_71439976 | 0.50 |
ENST00000536515.5
ENST00000540815.2 |
LGR5
|
leucine rich repeat containing G protein-coupled receptor 5 |
| chr19_-_2042066 | 0.50 |
ENST00000591588.1
ENST00000591142.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr6_+_128883114 | 0.50 |
ENST00000421865.3
ENST00000618192.4 ENST00000617695.4 |
LAMA2
|
laminin subunit alpha 2 |
| chr11_-_65058521 | 0.50 |
ENST00000340252.8
ENST00000355721.7 ENST00000356632.7 ENST00000358658.8 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase like 1 |
| chr7_-_143647646 | 0.50 |
ENST00000636941.1
|
TCAF2C
|
TRPM8 channel associated factor 2C |
| chr7_-_102543849 | 0.49 |
ENST00000644544.1
|
UPK3BL2
|
uroplakin 3B like 2 |
| chr16_-_74607088 | 0.49 |
ENST00000565260.1
ENST00000447066.6 ENST00000205061.9 ENST00000422840.7 ENST00000627032.2 |
GLG1
|
golgi glycoprotein 1 |
| chr4_-_41214602 | 0.49 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr19_-_41428730 | 0.49 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chrX_-_47629845 | 0.49 |
ENST00000469388.1
ENST00000396992.8 ENST00000377005.6 |
CFP
|
complement factor properdin |
| chr8_-_61646807 | 0.48 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr2_+_10123171 | 0.48 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr16_-_90019414 | 0.48 |
ENST00000002501.11
|
DBNDD1
|
dysbindin domain containing 1 |
| chr17_-_41812586 | 0.48 |
ENST00000355468.7
ENST00000590496.1 |
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr2_+_79512993 | 0.48 |
ENST00000496558.5
ENST00000451966.5 ENST00000402739.9 ENST00000629316.2 |
CTNNA2
|
catenin alpha 2 |
| chr6_-_65707214 | 0.48 |
ENST00000370621.7
ENST00000393380.6 ENST00000503581.6 |
EYS
|
eyes shut homolog |
| chr9_-_23821275 | 0.47 |
ENST00000380110.8
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr12_+_8992029 | 0.47 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.8 | 2.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.7 | 2.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 2.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.6 | 1.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 3.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.5 | 7.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.5 | 0.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.4 | 3.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 1.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.3 | 1.4 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.3 | 1.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 0.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 4.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.3 | 0.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 1.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.0 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.2 | 1.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.7 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.2 | 0.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.6 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.2 | 1.1 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 1.0 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 1.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.8 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.6 | GO:0008355 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.2 | 0.7 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.6 | GO:0051714 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 0.5 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 1.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 0.8 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.6 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 1.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.9 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.4 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 1.8 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 2.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 25.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 1.0 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.0 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.7 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 2.8 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 1.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.7 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.6 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 1.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.6 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) |
| 0.1 | 0.3 | GO:0098758 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.3 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.8 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 1.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:2000077 | negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.5 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.2 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.6 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.2 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.4 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.3 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) B cell selection(GO:0002339) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.4 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.6 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.3 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.1 | 1.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 1.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.5 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.9 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.2 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.2 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.6 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.4 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.9 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.3 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.9 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:1904782 | negative regulation of glutamate receptor signaling pathway(GO:1900450) negative regulation of NMDA glutamate receptor activity(GO:1904782) |
| 0.0 | 1.3 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.9 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.2 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.9 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.8 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 4.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.2 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 1.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0060932 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 0.0 | 0.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.7 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0061643 | vagus nerve morphogenesis(GO:0021644) chemorepulsion of branchiomotor axon(GO:0021793) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 1.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.9 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of odontogenesis(GO:0042482) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 2.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.4 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.4 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.0 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.3 | GO:0040032 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 1.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 1.0 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.8 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.5 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 1.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.3 | 1.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 0.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.7 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 2.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 14.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.6 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.8 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 2.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 4.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 5.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 3.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 6.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 1.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 2.7 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.7 | 1.4 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.6 | 2.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.4 | 3.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 1.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 2.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 3.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 0.9 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 1.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 2.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.5 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.3 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 1.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.5 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 3.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0009032 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 1.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.4 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 11.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 2.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 1.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 9.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 8.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 3.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |