Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
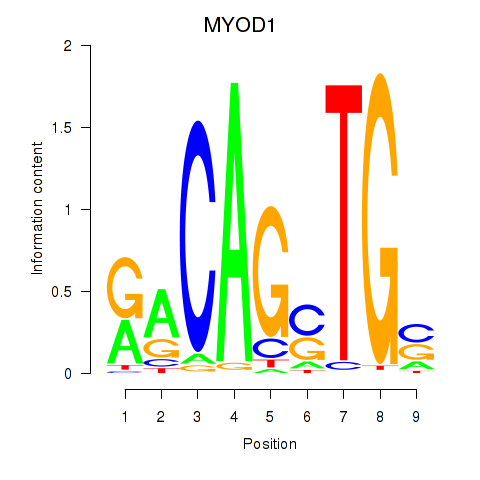
Results for MYOD1
Z-value: 0.83
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.4 | MYOD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYOD1 | hg38_v1_chr11_+_17719564_17719577 | -0.19 | 3.1e-01 | Click! |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 1.0 | 2.9 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.9 | 2.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.8 | 2.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.6 | 1.7 | GO:0090031 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) positive regulation of steroid hormone biosynthetic process(GO:0090031) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.5 | 2.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.5 | 1.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 1.2 | GO:0071657 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.4 | 1.5 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.4 | 4.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.3 | 2.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 1.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.3 | 1.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.3 | 1.4 | GO:0097195 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.3 | 0.8 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 0.7 | GO:2000182 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.7 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.2 | 1.7 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 3.8 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.2 | 0.6 | GO:0010933 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.2 | 4.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.2 | 2.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 0.5 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 1.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 0.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 0.7 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.5 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.2 | 1.0 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.2 | 0.6 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.9 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 1.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.4 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 1.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 1.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 1.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.8 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.5 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 2.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.4 | GO:1903891 | regulation of ATF6-mediated unfolded protein response(GO:1903891) |
| 0.1 | 0.5 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0034031 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.1 | 0.3 | GO:0003175 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.3 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.2 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) Wnt signaling pathway involved in somitogenesis(GO:0090244) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.7 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:1900154 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 1.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.8 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.5 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.1 | 2.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.2 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 5.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 0.8 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0099624 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.8 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 2.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 1.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.4 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 1.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.8 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.6 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.7 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 1.7 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.4 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.5 | GO:0048208 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.0 | GO:0034552 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 6.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 2.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 1.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 1.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 0.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 1.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 3.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 2.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 3.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 2.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 2.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.3 | 3.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 2.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.7 | 2.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 0.9 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 0.8 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.2 | 0.7 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.2 | 2.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 2.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.7 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.7 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.2 | 1.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 1.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 6.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 1.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 3.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 3.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.4 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 4.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.9 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |