Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
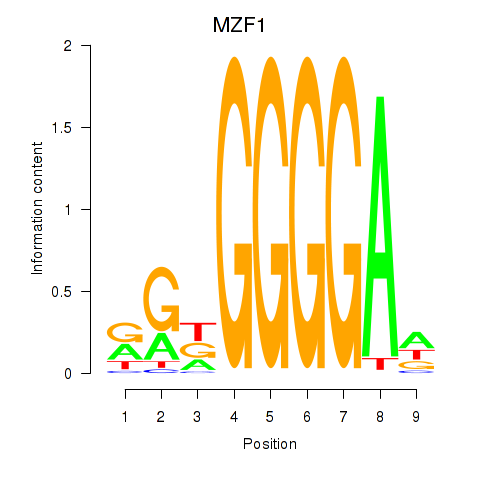
Results for MZF1
Z-value: 1.01
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.9 | MZF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg38_v1_chr19_-_58573555_58573582, hg38_v1_chr19_-_58573280_58573354 | -0.37 | 4.3e-02 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.6 | 1.9 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.5 | 1.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.5 | 1.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.5 | 1.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.5 | 1.5 | GO:0007315 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.5 | 2.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.4 | 1.2 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 0.4 | 1.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.4 | 2.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 2.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.0 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.3 | 1.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 1.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.3 | 1.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 2.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.3 | 1.9 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 1.7 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.6 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.3 | 1.9 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.8 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 1.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 1.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 0.7 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.7 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.9 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.2 | 1.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 3.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 0.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 0.7 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 4.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.6 | GO:1902688 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) |
| 0.2 | 0.6 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.2 | 1.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.2 | 2.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 0.8 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.9 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 2.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 | 0.7 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 4.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.7 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.2 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 1.8 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.8 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 1.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 | 2.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 2.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.6 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.5 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 1.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.6 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.5 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.4 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 2.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 2.5 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.9 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:0090133 | corticotropin hormone secreting cell differentiation(GO:0060128) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 6.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.4 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.7 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.5 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.3 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 4.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.2 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 1.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.4 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.5 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.8 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 1.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.6 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.4 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 0.2 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.1 | 0.2 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.1 | GO:1905006 | negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 | 0.4 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.5 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.2 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.0 | 0.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.5 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.5 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.2 | GO:0046668 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) regulation of retinal cell programmed cell death(GO:0046668) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.0 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.6 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 1.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 1.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.9 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 1.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.0 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.9 | GO:0090207 | regulation of triglyceride metabolic process(GO:0090207) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 1.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.2 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0098907 | protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.5 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1902513 | neurofilament bundle assembly(GO:0033693) regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 1.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.0 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.6 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.4 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.0 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.0 | 0.1 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.3 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 5.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.0 | GO:1904328 | lymphatic endothelial cell fate commitment(GO:0060838) positive regulation of platelet aggregation(GO:1901731) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 1.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.0 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.6 | 2.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 4.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 2.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 2.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 2.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.3 | 5.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.3 | 2.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 1.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 1.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.6 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 1.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 0.5 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 1.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 3.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 1.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 2.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 2.0 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 2.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 1.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.2 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 1.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 1.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 6.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0030395 | lactose binding(GO:0030395) |
| 0.5 | 1.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 2.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.9 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 1.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 2.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.9 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.2 | 1.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 0.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.2 | 0.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 0.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.2 | 0.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 0.3 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 3.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.2 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 1.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.5 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 1.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.0 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 1.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 2.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.7 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 1.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 6.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.1 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 2.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.5 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.7 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 1.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 1.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 4.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 5.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 6.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 5.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 13.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 2.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |