Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
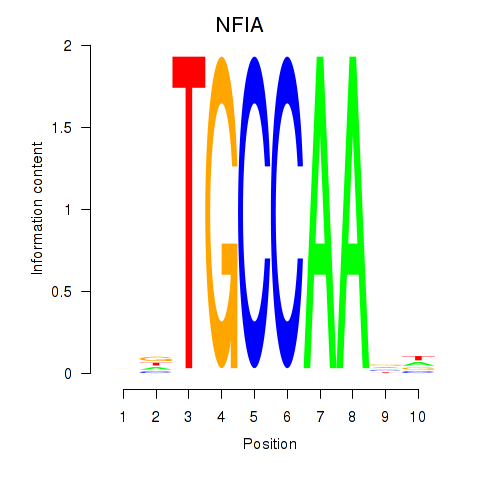
Results for NFIA
Z-value: 1.08
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.17 | NFIA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg38_v1_chr1_+_61404076_61404118 | -0.77 | 6.7e-07 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_41703062 | 7.73 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr2_-_31414694 | 7.58 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr13_-_20230970 | 4.39 |
ENST00000644667.1
ENST00000646108.1 |
GJB6
|
gap junction protein beta 6 |
| chr21_-_26843012 | 3.81 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 3.71 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr4_-_56681588 | 3.35 |
ENST00000554144.5
ENST00000381260.7 |
HOPX
|
HOP homeobox |
| chr4_-_56681288 | 3.33 |
ENST00000556376.6
ENST00000420433.6 |
HOPX
|
HOP homeobox |
| chr10_+_100347225 | 2.99 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase |
| chrX_+_136169624 | 2.97 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_136169833 | 2.93 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr2_-_112784486 | 2.87 |
ENST00000263339.4
|
IL1A
|
interleukin 1 alpha |
| chr5_+_136059151 | 2.78 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor beta induced |
| chr15_+_67166019 | 2.75 |
ENST00000537194.6
|
SMAD3
|
SMAD family member 3 |
| chrX_+_136169664 | 2.66 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr5_-_95682968 | 2.64 |
ENST00000274432.13
|
SPATA9
|
spermatogenesis associated 9 |
| chr13_+_77535681 | 2.60 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 2.58 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr13_+_77535669 | 2.57 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr10_-_99235783 | 2.50 |
ENST00000370546.5
ENST00000614306.1 |
HPSE2
|
heparanase 2 (inactive) |
| chr7_-_27095972 | 2.48 |
ENST00000355633.5
ENST00000643460.2 |
HOXA1
|
homeobox A1 |
| chr12_-_121800558 | 2.44 |
ENST00000546227.5
|
RHOF
|
ras homolog family member F, filopodia associated |
| chr12_-_91111460 | 2.37 |
ENST00000266718.5
|
LUM
|
lumican |
| chr9_-_96302142 | 2.35 |
ENST00000648799.1
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr3_-_74521140 | 2.30 |
ENST00000263665.6
|
CNTN3
|
contactin 3 |
| chr9_-_96302104 | 2.24 |
ENST00000375262.4
ENST00000650386.1 |
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr2_+_102337148 | 2.20 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr19_+_41219177 | 2.17 |
ENST00000301178.9
|
AXL
|
AXL receptor tyrosine kinase |
| chrX_+_136169891 | 2.15 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr14_+_75279637 | 2.09 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr8_+_123182635 | 2.08 |
ENST00000276699.10
ENST00000522648.5 |
FAM83A
|
family with sequence similarity 83 member A |
| chr14_+_75280078 | 2.04 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr14_-_74612226 | 1.90 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr11_-_128522264 | 1.87 |
ENST00000531611.5
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr18_+_63587297 | 1.86 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr7_+_76424922 | 1.83 |
ENST00000394857.8
|
ZP3
|
zona pellucida glycoprotein 3 |
| chr11_-_128522189 | 1.79 |
ENST00000526145.6
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr18_+_63587336 | 1.78 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr14_-_24263162 | 1.75 |
ENST00000206765.11
ENST00000544573.5 |
TGM1
|
transglutaminase 1 |
| chr9_-_96302170 | 1.74 |
ENST00000375263.8
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr3_+_52316319 | 1.68 |
ENST00000420323.7
|
DNAH1
|
dynein axonemal heavy chain 1 |
| chr16_-_55833186 | 1.66 |
ENST00000361503.8
ENST00000422046.6 |
CES1
|
carboxylesterase 1 |
| chr11_-_58575846 | 1.62 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr15_+_41256907 | 1.62 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr11_-_128522285 | 1.60 |
ENST00000319397.6
ENST00000535549.5 |
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr18_+_63476927 | 1.60 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr12_-_95116967 | 1.59 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_-_43313473 | 1.55 |
ENST00000433297.2
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr14_-_63728027 | 1.55 |
ENST00000247225.7
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr4_-_102345196 | 1.54 |
ENST00000683412.1
ENST00000682227.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr3_+_172754457 | 1.54 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr20_+_6767678 | 1.53 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr1_+_209704836 | 1.52 |
ENST00000367027.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr5_-_43313403 | 1.50 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr22_-_38302990 | 1.47 |
ENST00000612795.2
ENST00000451964.5 |
CSNK1E
|
casein kinase 1 epsilon |
| chr12_+_57434778 | 1.39 |
ENST00000309668.3
|
INHBC
|
inhibin subunit beta C |
| chr15_+_66453418 | 1.38 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr2_+_17539964 | 1.35 |
ENST00000457525.5
|
VSNL1
|
visinin like 1 |
| chr21_-_43427131 | 1.35 |
ENST00000270162.8
|
SIK1
|
salt inducible kinase 1 |
| chr11_+_33016106 | 1.33 |
ENST00000311388.7
|
DEPDC7
|
DEP domain containing 7 |
| chr15_+_67125707 | 1.31 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr21_+_6111123 | 1.31 |
ENST00000613488.3
|
SIK1B
|
salt inducible kinase 1B (putative) |
| chr18_+_58862904 | 1.26 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532 |
| chr1_-_24143112 | 1.25 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr12_-_119803383 | 1.23 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr5_+_93584916 | 1.23 |
ENST00000647447.1
ENST00000615873.1 |
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr1_+_24319511 | 1.21 |
ENST00000356046.6
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr1_-_208244375 | 1.21 |
ENST00000367033.4
|
PLXNA2
|
plexin A2 |
| chr4_-_102345469 | 1.21 |
ENST00000356736.5
ENST00000682932.1 |
SLC39A8
|
solute carrier family 39 member 8 |
| chr12_-_27972725 | 1.17 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr4_-_156970903 | 1.16 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_+_44847874 | 1.16 |
ENST00000253410.3
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr10_+_5196831 | 1.15 |
ENST00000263126.3
|
AKR1C4
|
aldo-keto reductase family 1 member C4 |
| chr10_-_99235846 | 1.15 |
ENST00000370552.8
ENST00000370549.5 ENST00000628193.2 |
HPSE2
|
heparanase 2 (inactive) |
| chr2_+_135531460 | 1.14 |
ENST00000683871.1
ENST00000409478.5 ENST00000264160.8 ENST00000438014.5 |
R3HDM1
|
R3H domain containing 1 |
| chr1_-_120051714 | 1.13 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr17_-_8210203 | 1.13 |
ENST00000578549.5
ENST00000582368.5 |
AURKB
|
aurora kinase B |
| chr12_-_52814106 | 1.13 |
ENST00000551956.2
|
KRT4
|
keratin 4 |
| chr1_-_151008365 | 1.12 |
ENST00000361936.9
ENST00000361738.11 |
MINDY1
|
MINDY lysine 48 deubiquitinase 1 |
| chr3_+_57890011 | 1.11 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr14_+_51847145 | 1.11 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr16_-_79600727 | 1.09 |
ENST00000326043.5
|
MAF
|
MAF bZIP transcription factor |
| chr19_-_43465596 | 1.08 |
ENST00000244333.4
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr18_+_31591869 | 1.08 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr4_+_94489030 | 1.08 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chrX_+_12906639 | 1.08 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr3_+_50269140 | 1.07 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr15_+_41286011 | 1.07 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr7_-_27143672 | 1.07 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr5_-_78549151 | 1.05 |
ENST00000515007.6
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr17_-_74361860 | 1.05 |
ENST00000375366.4
|
BTBD17
|
BTB domain containing 17 |
| chr2_-_65432591 | 1.04 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr8_-_130386864 | 1.01 |
ENST00000521426.5
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr7_-_151633182 | 1.01 |
ENST00000476632.2
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr19_-_39834127 | 1.01 |
ENST00000601972.1
ENST00000430012.6 ENST00000323039.10 ENST00000348817.7 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr12_+_93571832 | 1.00 |
ENST00000549887.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr19_-_39833615 | 0.99 |
ENST00000593685.5
ENST00000600611.5 |
DYRK1B
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr7_-_23347704 | 0.97 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr10_+_133527355 | 0.97 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr6_-_30690968 | 0.95 |
ENST00000376420.9
ENST00000376421.7 |
NRM
|
nurim |
| chr4_+_143433491 | 0.95 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr10_+_103245887 | 0.94 |
ENST00000441178.2
|
RPEL1
|
ribulose-5-phosphate-3-epimerase like 1 |
| chr11_-_2149603 | 0.93 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein |
| chr12_-_98644733 | 0.92 |
ENST00000299157.5
ENST00000393042.3 |
IKBIP
|
IKBKB interacting protein |
| chr9_-_69672341 | 0.91 |
ENST00000265381.7
|
APBA1
|
amyloid beta precursor protein binding family A member 1 |
| chr12_+_113244261 | 0.90 |
ENST00000392569.8
ENST00000552542.5 |
TPCN1
|
two pore segment channel 1 |
| chr17_-_40501615 | 0.90 |
ENST00000254051.11
|
TNS4
|
tensin 4 |
| chr3_+_101779182 | 0.90 |
ENST00000495842.5
ENST00000273347.10 |
NXPE3
|
neurexophilin and PC-esterase domain family member 3 |
| chr2_-_89085787 | 0.89 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr3_+_111999326 | 0.89 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chrX_-_132219473 | 0.89 |
ENST00000620646.4
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr17_-_41505597 | 0.88 |
ENST00000336861.7
ENST00000246635.8 ENST00000587544.5 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr5_-_88785493 | 0.88 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr10_-_124450027 | 0.88 |
ENST00000451024.5
|
NKX1-2
|
NK1 homeobox 2 |
| chr3_-_37174578 | 0.88 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr16_+_8797813 | 0.87 |
ENST00000268261.9
ENST00000569958.5 |
PMM2
|
phosphomannomutase 2 |
| chr13_-_39603123 | 0.87 |
ENST00000379589.4
|
LHFPL6
|
LHFPL tetraspan subfamily member 6 |
| chrX_-_132219439 | 0.87 |
ENST00000370874.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr15_+_73873604 | 0.86 |
ENST00000535547.6
ENST00000562056.1 |
TBC1D21
|
TBC1 domain family member 21 |
| chr8_-_28386417 | 0.85 |
ENST00000521185.5
ENST00000520290.5 ENST00000344423.10 |
ZNF395
|
zinc finger protein 395 |
| chr1_-_155254908 | 0.84 |
ENST00000491082.1
ENST00000350210.6 ENST00000368368.7 |
FAM189B
|
family with sequence similarity 189 member B |
| chr6_+_43021606 | 0.84 |
ENST00000244496.6
|
RRP36
|
ribosomal RNA processing 36 |
| chr7_+_129375643 | 0.84 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr19_-_13116649 | 0.84 |
ENST00000437766.5
ENST00000221504.12 |
TRMT1
|
tRNA methyltransferase 1 |
| chr4_+_37960397 | 0.83 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr10_+_24239181 | 0.83 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217 |
| chr12_-_52777343 | 0.82 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr19_-_13116719 | 0.82 |
ENST00000588229.1
ENST00000357720.9 |
TRMT1
|
tRNA methyltransferase 1 |
| chr20_+_44337043 | 0.82 |
ENST00000217043.4
|
R3HDML
|
R3H domain containing like |
| chr5_+_148268830 | 0.81 |
ENST00000511106.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr4_-_76007501 | 0.81 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr8_+_38728550 | 0.81 |
ENST00000520340.5
ENST00000518415.5 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
| chr12_+_93570969 | 0.80 |
ENST00000536696.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr3_-_47892743 | 0.78 |
ENST00000420772.6
|
MAP4
|
microtubule associated protein 4 |
| chr19_-_45792755 | 0.77 |
ENST00000377735.7
ENST00000270223.7 |
DMWD
|
DM1 locus, WD repeat containing |
| chr6_-_132757883 | 0.75 |
ENST00000525289.5
ENST00000326499.11 |
VNN2
|
vanin 2 |
| chr11_-_2137277 | 0.74 |
ENST00000381392.5
ENST00000381395.5 ENST00000418738.2 |
IGF2
|
insulin like growth factor 2 |
| chr14_-_59870752 | 0.73 |
ENST00000611068.1
ENST00000267484.10 |
RTN1
|
reticulon 1 |
| chr17_+_19411220 | 0.73 |
ENST00000461366.2
|
RNF112
|
ring finger protein 112 |
| chr2_-_227164194 | 0.72 |
ENST00000396625.5
|
COL4A4
|
collagen type IV alpha 4 chain |
| chr11_+_34632464 | 0.72 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr10_+_72893734 | 0.72 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr4_+_2818155 | 0.71 |
ENST00000511747.6
|
SH3BP2
|
SH3 domain binding protein 2 |
| chr2_-_24328113 | 0.70 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr10_+_11164961 | 0.70 |
ENST00000399850.7
ENST00000417956.6 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr19_+_40751179 | 0.70 |
ENST00000243563.8
ENST00000601393.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr17_-_41612757 | 0.70 |
ENST00000301653.9
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr21_-_5973383 | 0.70 |
ENST00000464664.3
|
ENSG00000274559.3
|
novel histone H2B family protein |
| chr12_+_56128217 | 0.69 |
ENST00000267113.4
ENST00000394048.10 |
ESYT1
|
extended synaptotagmin 1 |
| chr12_+_56041893 | 0.69 |
ENST00000552361.1
ENST00000646449.2 |
RPS26
|
ribosomal protein S26 |
| chr10_-_73433550 | 0.69 |
ENST00000299432.7
|
MSS51
|
MSS51 mitochondrial translational activator |
| chrX_+_12906612 | 0.68 |
ENST00000218032.7
|
TLR8
|
toll like receptor 8 |
| chr6_-_31902041 | 0.68 |
ENST00000375527.3
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr17_+_42017020 | 0.68 |
ENST00000307641.9
|
NKIRAS2
|
NFKB inhibitor interacting Ras like 2 |
| chr6_-_155455830 | 0.67 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3 |
| chr3_-_171460063 | 0.67 |
ENST00000284483.12
ENST00000475336.5 ENST00000357327.9 ENST00000460047.5 ENST00000488470.5 ENST00000470834.5 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr3_+_152300135 | 0.66 |
ENST00000465907.6
ENST00000492948.5 ENST00000485509.5 ENST00000464596.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chrX_+_50067576 | 0.66 |
ENST00000376108.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr6_+_99606833 | 0.66 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13 |
| chr18_-_63158208 | 0.65 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr19_-_40690629 | 0.65 |
ENST00000252891.8
|
NUMBL
|
NUMB like endocytic adaptor protein |
| chr10_+_72273914 | 0.65 |
ENST00000681898.1
ENST00000307365.4 |
DDIT4
|
DNA damage inducible transcript 4 |
| chr20_-_14337602 | 0.65 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr8_-_143939543 | 0.64 |
ENST00000345136.8
|
PLEC
|
plectin |
| chr17_+_16217198 | 0.64 |
ENST00000581006.5
ENST00000584797.5 ENST00000225609.10 ENST00000395844.8 ENST00000463810.2 |
PIGL
|
phosphatidylinositol glycan anchor biosynthesis class L |
| chr4_-_102345061 | 0.64 |
ENST00000394833.6
|
SLC39A8
|
solute carrier family 39 member 8 |
| chr5_-_55712280 | 0.64 |
ENST00000506624.5
ENST00000513275.5 ENST00000513993.5 ENST00000396865.7 ENST00000503891.5 ENST00000507109.5 |
SLC38A9
|
solute carrier family 38 member 9 |
| chr1_-_237004440 | 0.63 |
ENST00000464121.3
|
MT1HL1
|
metallothionein 1H like 1 |
| chr3_+_19148500 | 0.63 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr4_+_73404255 | 0.62 |
ENST00000621628.4
ENST00000621085.4 ENST00000415165.6 ENST00000295897.9 ENST00000503124.5 ENST00000509063.5 ENST00000401494.7 |
ALB
|
albumin |
| chr6_+_116399395 | 0.62 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr16_-_19714577 | 0.62 |
ENST00000567367.1
|
KNOP1
|
lysine rich nucleolar protein 1 |
| chr19_-_1822038 | 0.61 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog |
| chr7_-_84194781 | 0.61 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A |
| chr17_-_39864140 | 0.61 |
ENST00000623724.3
ENST00000439167.6 ENST00000377945.7 ENST00000394189.6 ENST00000377944.7 ENST00000377958.6 ENST00000535189.5 ENST00000377952.6 |
IKZF3
|
IKAROS family zinc finger 3 |
| chr1_+_43933794 | 0.61 |
ENST00000372359.10
ENST00000498139.6 ENST00000491846.5 |
ARTN
|
artemin |
| chr2_+_33476640 | 0.60 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr4_+_92303946 | 0.60 |
ENST00000282020.9
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr16_-_79600698 | 0.60 |
ENST00000393350.1
|
MAF
|
MAF bZIP transcription factor |
| chr3_-_134651011 | 0.60 |
ENST00000508956.5
ENST00000503669.1 ENST00000423778.7 |
KY
|
kyphoscoliosis peptidase |
| chr1_+_67685342 | 0.59 |
ENST00000617962.2
|
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr9_+_27109135 | 0.58 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr2_-_162243375 | 0.58 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr12_+_93572664 | 0.58 |
ENST00000551556.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr1_+_67685170 | 0.58 |
ENST00000370985.4
ENST00000370986.9 ENST00000650283.1 ENST00000648742.1 |
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr14_+_22829879 | 0.58 |
ENST00000355151.9
ENST00000397496.7 ENST00000555345.5 ENST00000432849.7 ENST00000553711.5 ENST00000556465.5 ENST00000397505.2 ENST00000557221.1 ENST00000556840.5 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr17_-_40937445 | 0.57 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr5_+_148268741 | 0.57 |
ENST00000398450.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr6_-_32816910 | 0.57 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr8_+_32721823 | 0.56 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr17_-_40937641 | 0.56 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr4_+_155903688 | 0.56 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_-_1485733 | 0.56 |
ENST00000648446.1
|
MYO1C
|
myosin IC |
| chr9_+_100442271 | 0.56 |
ENST00000502978.1
|
MSANTD3-TMEFF1
|
MSANTD3-TMEFF1 readthrough |
| chrX_-_120559889 | 0.55 |
ENST00000371323.3
|
CUL4B
|
cullin 4B |
| chr11_+_126355894 | 0.55 |
ENST00000530591.5
ENST00000534083.5 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr17_+_7420315 | 0.55 |
ENST00000323675.4
|
SPEM1
|
spermatid maturation 1 |
| chr7_+_95485898 | 0.55 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_+_148133684 | 0.55 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr11_+_10305065 | 0.55 |
ENST00000534464.1
ENST00000278175.10 ENST00000530439.1 ENST00000524948.5 ENST00000528655.5 ENST00000526492.4 ENST00000525063.2 |
ADM
|
adrenomedullin |
| chr12_+_7155867 | 0.55 |
ENST00000535313.2
ENST00000331148.5 |
CLSTN3
|
calsyntenin 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 2.5 | 7.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.7 | 4.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.7 | 6.7 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.5 | 1.5 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.5 | 5.3 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.5 | 3.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.5 | 4.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.4 | 1.6 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.4 | 7.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.4 | 1.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.4 | 2.8 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.4 | 1.2 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.4 | 2.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.4 | 1.8 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.3 | 3.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 1.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 1.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.3 | 2.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.3 | 3.7 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.3 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 0.8 | GO:1904640 | response to methionine(GO:1904640) |
| 0.3 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 1.3 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.3 | 2.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.3 | 6.7 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 1.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.2 | 1.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 2.9 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 0.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 0.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.2 | 2.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 1.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 1.0 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.4 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 1.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.2 | 0.5 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.7 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.2 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.5 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.2 | 0.5 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.7 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 0.5 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.4 | GO:0060585 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 1.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.4 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 11.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.9 | GO:0019323 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.1 | 0.5 | GO:0015855 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.4 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.6 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.4 | GO:1903781 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of cardiac conduction(GO:1903781) |
| 0.1 | 0.6 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.3 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.8 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 1.0 | GO:0070120 | brainstem development(GO:0003360) ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 1.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 1.5 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.2 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 4.4 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.1 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:2000597 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.3 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.5 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 1.1 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.9 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.7 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.6 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 1.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.4 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 1.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.6 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.8 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 1.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:2000814 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 3.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 1.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.6 | GO:2001212 | regulation of vasculogenesis(GO:2001212) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 2.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0009631 | cold acclimation(GO:0009631) |
| 0.0 | 1.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 11.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 3.6 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.5 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 1.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 1.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) terpene metabolic process(GO:0042214) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 1.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 3.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 1.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.5 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 1.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0042946 | glucoside transport(GO:0042946) |
| 0.0 | 0.1 | GO:0060577 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 1.1 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.3 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) positive regulation of skeletal muscle fiber development(GO:0048743) pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.6 | GO:0042339 | keratan sulfate biosynthetic process(GO:0018146) keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.3 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.5 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 1.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.9 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.8 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 4.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.5 | 4.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 1.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.3 | 1.8 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.3 | 0.8 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.3 | 1.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 1.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 10.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 4.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.5 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 2.5 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 1.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 7.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 9.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 3.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 1.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 14.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 1.2 | 3.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.1 | 3.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.0 | 8.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.0 | 6.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.8 | 1.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.7 | 3.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.7 | 2.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.7 | 4.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.5 | 1.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.4 | 2.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.7 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 0.9 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.3 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 1.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 1.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 1.8 | GO:0032190 | manganese ion transmembrane transporter activity(GO:0005384) acrosin binding(GO:0032190) |
| 0.2 | 0.5 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.2 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 2.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 1.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 3.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 4.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.0 | GO:0008607 | AMP-activated protein kinase activity(GO:0004679) phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 1.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.5 | GO:0043208 | fucose binding(GO:0042806) glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 1.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 3.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 6.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 1.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 6.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 5.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 15.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 6.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 7.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 4.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 3.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 4.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.4 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |