Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
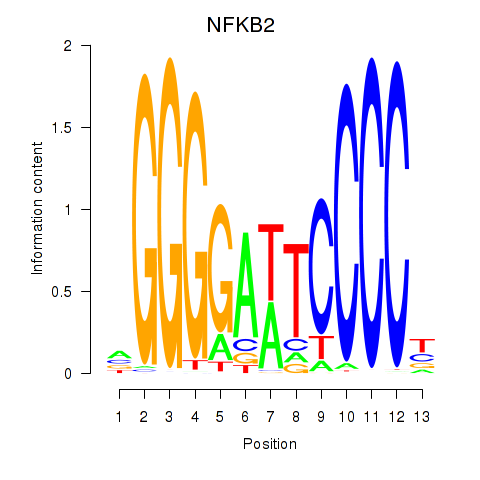
Results for NFKB2
Z-value: 0.75
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.20 | NFKB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg38_v1_chr10_+_102394488_102394540 | 0.20 | 3.0e-01 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_37244417 | 1.83 |
ENST00000405484.5
ENST00000441619.5 ENST00000406508.5 |
RAC2
|
Rac family small GTPase 2 |
| chr8_+_85463997 | 1.77 |
ENST00000285379.10
|
CA2
|
carbonic anhydrase 2 |
| chr22_-_37244237 | 1.72 |
ENST00000401529.3
ENST00000249071.11 |
RAC2
|
Rac family small GTPase 2 |
| chr4_-_73998669 | 1.45 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5 |
| chr3_+_111999326 | 1.40 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr19_+_2096960 | 1.36 |
ENST00000588003.5
|
IZUMO4
|
IZUMO family member 4 |
| chr4_+_17810945 | 1.33 |
ENST00000251496.7
|
NCAPG
|
non-SMC condensin I complex subunit G |
| chr1_-_186680411 | 1.31 |
ENST00000367468.10
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 |
| chr15_-_74725370 | 1.28 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr19_+_2097010 | 1.24 |
ENST00000610800.1
|
IZUMO4
|
IZUMO family member 4 |
| chr18_+_23949847 | 1.23 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr19_+_2096937 | 1.22 |
ENST00000395296.5
|
IZUMO4
|
IZUMO family member 4 |
| chrX_+_65667645 | 1.20 |
ENST00000360270.7
|
MSN
|
moesin |
| chr8_+_53880894 | 1.20 |
ENST00000276500.4
|
RGS20
|
regulator of G protein signaling 20 |
| chrX_+_103918872 | 1.19 |
ENST00000419165.5
|
TMSB15B
|
thymosin beta 15B |
| chr8_+_53880867 | 1.17 |
ENST00000522225.5
|
RGS20
|
regulator of G protein signaling 20 |
| chr21_-_43659460 | 1.15 |
ENST00000443485.1
ENST00000291560.7 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr19_+_48321454 | 1.15 |
ENST00000599704.5
|
EMP3
|
epithelial membrane protein 3 |
| chrX_-_154366334 | 1.14 |
ENST00000673639.2
|
FLNA
|
filamin A |
| chr12_-_57767057 | 1.12 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr12_+_53097656 | 1.08 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr15_+_85380625 | 1.05 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr3_+_111998739 | 0.98 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111999189 | 0.96 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr11_-_66907891 | 0.96 |
ENST00000393955.6
|
PC
|
pyruvate carboxylase |
| chr1_-_26360050 | 0.94 |
ENST00000475866.3
|
CRYBG2
|
crystallin beta-gamma domain containing 2 |
| chr3_+_111998915 | 0.92 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr1_-_37634864 | 0.89 |
ENST00000356545.7
|
RSPO1
|
R-spondin 1 |
| chr10_+_110497898 | 0.88 |
ENST00000369583.4
|
DUSP5
|
dual specificity phosphatase 5 |
| chr19_+_53867874 | 0.86 |
ENST00000448420.5
ENST00000439000.5 ENST00000391771.1 ENST00000391770.9 |
MYADM
|
myeloid associated differentiation marker |
| chr22_-_38317380 | 0.83 |
ENST00000413574.6
|
CSNK1E
|
casein kinase 1 epsilon |
| chr16_-_74700786 | 0.82 |
ENST00000306247.11
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr2_+_64454506 | 0.82 |
ENST00000409537.2
|
LGALSL
|
galectin like |
| chrX_-_15854743 | 0.78 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_-_6485433 | 0.76 |
ENST00000535355.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr20_+_45174894 | 0.75 |
ENST00000243924.4
|
PI3
|
peptidase inhibitor 3 |
| chr5_-_138331770 | 0.75 |
ENST00000415130.6
ENST00000323760.11 ENST00000503022.5 |
CDC25C
|
cell division cycle 25C |
| chr3_-_50303565 | 0.74 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr1_-_112956063 | 0.73 |
ENST00000538576.5
ENST00000369626.8 ENST00000458229.6 |
SLC16A1
|
solute carrier family 16 member 1 |
| chr16_-_74700845 | 0.72 |
ENST00000308807.12
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr2_-_89010515 | 0.70 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr10_+_92591733 | 0.70 |
ENST00000676647.1
|
KIF11
|
kinesin family member 11 |
| chr12_+_109573757 | 0.67 |
ENST00000228510.8
ENST00000539696.5 ENST00000392727.7 |
MVK
|
mevalonate kinase |
| chr22_+_43923755 | 0.66 |
ENST00000423180.2
ENST00000216180.8 |
PNPLA3
|
patatin like phospholipase domain containing 3 |
| chr7_+_151114597 | 0.64 |
ENST00000335367.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_208244375 | 0.63 |
ENST00000367033.4
|
PLXNA2
|
plexin A2 |
| chr2_-_88947820 | 0.62 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr12_-_54419259 | 0.62 |
ENST00000293379.9
|
ITGA5
|
integrin subunit alpha 5 |
| chr4_-_20984011 | 0.61 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr19_-_1513189 | 0.60 |
ENST00000395467.6
|
ADAMTSL5
|
ADAMTS like 5 |
| chr3_-_108589375 | 0.60 |
ENST00000625495.1
ENST00000619684.4 ENST00000295746.13 |
CIP2A
|
cellular inhibitor of PP2A |
| chr11_+_61792878 | 0.59 |
ENST00000305885.3
ENST00000535723.1 |
FEN1
|
flap structure-specific endonuclease 1 |
| chr19_+_44643798 | 0.59 |
ENST00000344956.8
|
PVR
|
PVR cell adhesion molecule |
| chr22_+_35400115 | 0.57 |
ENST00000382011.9
ENST00000216122.9 ENST00000416905.1 |
MCM5
|
minichromosome maintenance complex component 5 |
| chr1_+_111227610 | 0.56 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2 |
| chr19_-_4831689 | 0.56 |
ENST00000248244.6
|
TICAM1
|
toll like receptor adaptor molecule 1 |
| chr11_+_67056805 | 0.55 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D |
| chr12_+_6914571 | 0.55 |
ENST00000229277.6
ENST00000538763.5 ENST00000545045.6 |
ENO2
|
enolase 2 |
| chr1_-_209651291 | 0.54 |
ENST00000391911.5
ENST00000415782.1 |
LAMB3
|
laminin subunit beta 3 |
| chr1_+_165895583 | 0.54 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_165895564 | 0.54 |
ENST00000469256.6
|
UCK2
|
uridine-cytidine kinase 2 |
| chr16_+_88638569 | 0.54 |
ENST00000244241.5
|
IL17C
|
interleukin 17C |
| chr16_-_87869497 | 0.54 |
ENST00000261622.5
|
SLC7A5
|
solute carrier family 7 member 5 |
| chr14_-_50830479 | 0.53 |
ENST00000382043.8
|
NIN
|
ninein |
| chr1_+_111227699 | 0.53 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2 |
| chr20_+_61599755 | 0.52 |
ENST00000543233.2
|
CDH4
|
cadherin 4 |
| chr17_-_5583957 | 0.51 |
ENST00000354411.7
ENST00000577119.5 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr3_-_197298558 | 0.51 |
ENST00000656944.1
ENST00000346964.6 ENST00000448528.6 ENST00000655488.1 ENST00000357674.9 ENST00000667157.1 ENST00000661336.1 ENST00000654737.1 ENST00000659716.1 ENST00000657381.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chrX_-_15854791 | 0.50 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_-_154970735 | 0.50 |
ENST00000368445.9
ENST00000448116.7 ENST00000368449.8 |
SHC1
|
SHC adaptor protein 1 |
| chr10_-_84241538 | 0.48 |
ENST00000372105.4
|
LRIT1
|
leucine rich repeat, Ig-like and transmembrane domains 1 |
| chrX_+_68829009 | 0.48 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr11_+_706595 | 0.48 |
ENST00000531348.5
ENST00000530636.5 |
EPS8L2
|
EPS8 like 2 |
| chr5_+_136058849 | 0.48 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr1_-_205775182 | 0.48 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family |
| chr12_+_112938523 | 0.48 |
ENST00000679483.1
ENST00000679493.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr12_+_112938284 | 0.47 |
ENST00000681346.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr20_-_45124463 | 0.47 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr7_-_1556194 | 0.46 |
ENST00000297477.10
|
TMEM184A
|
transmembrane protein 184A |
| chr17_+_4951080 | 0.46 |
ENST00000521811.5
ENST00000323997.10 ENST00000522249.5 ENST00000519584.5 ENST00000519602.6 |
ENO3
|
enolase 3 |
| chr11_+_706196 | 0.46 |
ENST00000534755.5
ENST00000650127.1 |
EPS8L2
|
EPS8 like 2 |
| chr1_-_205775449 | 0.46 |
ENST00000235932.8
ENST00000437324.6 ENST00000414729.1 ENST00000367139.8 |
RAB29
|
RAB29, member RAS oncogene family |
| chr8_-_9150648 | 0.45 |
ENST00000310455.4
|
PPP1R3B
|
protein phosphatase 1 regulatory subunit 3B |
| chr16_+_28824116 | 0.44 |
ENST00000568266.5
|
ATXN2L
|
ataxin 2 like |
| chr19_+_54451290 | 0.44 |
ENST00000610347.1
|
LENG8
|
leukocyte receptor cluster member 8 |
| chr11_-_64803152 | 0.43 |
ENST00000439069.5
ENST00000294066.7 ENST00000377350.7 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_-_6485941 | 0.43 |
ENST00000676287.1
ENST00000400913.6 |
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr5_-_78648718 | 0.43 |
ENST00000380345.7
|
LHFPL2
|
LHFPL tetraspan subfamily member 2 |
| chr14_+_51847145 | 0.43 |
ENST00000615906.4
|
GNG2
|
G protein subunit gamma 2 |
| chr19_-_51723968 | 0.42 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr8_-_144416481 | 0.42 |
ENST00000276833.9
|
SLC39A4
|
solute carrier family 39 member 4 |
| chr4_+_141636923 | 0.42 |
ENST00000529613.5
|
IL15
|
interleukin 15 |
| chr3_+_12287962 | 0.42 |
ENST00000643197.2
ENST00000644622.2 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr11_+_706222 | 0.41 |
ENST00000318562.13
ENST00000533500.5 |
EPS8L2
|
EPS8 like 2 |
| chr11_+_706117 | 0.41 |
ENST00000533256.5
ENST00000614442.4 |
EPS8L2
|
EPS8 like 2 |
| chr12_-_76878985 | 0.41 |
ENST00000547435.1
ENST00000552330.5 ENST00000311083.10 ENST00000546966.5 |
CSRP2
|
cysteine and glycine rich protein 2 |
| chr3_+_12287899 | 0.41 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr19_+_44643902 | 0.41 |
ENST00000403059.8
ENST00000425690.8 |
PVR
|
PVR cell adhesion molecule |
| chr1_-_231241090 | 0.40 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr11_+_119185469 | 0.40 |
ENST00000525131.5
ENST00000355547.10 ENST00000531114.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr1_-_151993822 | 0.40 |
ENST00000368811.8
|
S100A10
|
S100 calcium binding protein A10 |
| chr3_+_12287859 | 0.40 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr2_-_223838022 | 0.40 |
ENST00000444408.1
|
AP1S3
|
adaptor related protein complex 1 subunit sigma 3 |
| chr7_+_142332182 | 0.40 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr2_-_218292496 | 0.40 |
ENST00000258412.8
ENST00000445635.5 ENST00000413976.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr11_-_18248632 | 0.40 |
ENST00000524555.3
ENST00000528349.5 ENST00000526900.1 ENST00000529528.5 ENST00000414546.6 |
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough serum amyloid A2 |
| chr19_+_44644025 | 0.39 |
ENST00000406449.8
|
PVR
|
PVR cell adhesion molecule |
| chr17_-_7205116 | 0.39 |
ENST00000649520.1
ENST00000649186.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr2_-_223837484 | 0.39 |
ENST00000446015.6
ENST00000409375.1 |
AP1S3
|
adaptor related protein complex 1 subunit sigma 3 |
| chr6_+_32637419 | 0.38 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr7_+_143316105 | 0.38 |
ENST00000343257.7
ENST00000650516.1 |
CLCN1
|
chloride voltage-gated channel 1 |
| chr1_-_147172456 | 0.38 |
ENST00000254101.4
|
PRKAB2
|
protein kinase AMP-activated non-catalytic subunit beta 2 |
| chr19_-_49896868 | 0.38 |
ENST00000593956.5
ENST00000391826.7 |
IL4I1
|
interleukin 4 induced 1 |
| chr1_-_41662298 | 0.38 |
ENST00000643665.1
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr19_+_1407731 | 0.38 |
ENST00000592453.2
|
DAZAP1
|
DAZ associated protein 1 |
| chr3_+_183253795 | 0.37 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr19_-_45423481 | 0.37 |
ENST00000589381.5
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr1_-_159945596 | 0.37 |
ENST00000361509.7
ENST00000611023.1 ENST00000368094.6 |
IGSF9
|
immunoglobulin superfamily member 9 |
| chr11_+_65919480 | 0.37 |
ENST00000527119.5
|
DRAP1
|
DR1 associated protein 1 |
| chr19_-_3786254 | 0.37 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr2_+_173075714 | 0.36 |
ENST00000409176.6
ENST00000338983.7 ENST00000375213.8 |
MAP3K20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr11_+_65919383 | 0.36 |
ENST00000312515.7
|
DRAP1
|
DR1 associated protein 1 |
| chr1_-_27356471 | 0.36 |
ENST00000486046.5
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr10_-_131981948 | 0.36 |
ENST00000633835.1
|
BNIP3
|
BCL2 interacting protein 3 |
| chr11_+_35618450 | 0.36 |
ENST00000317811.6
|
FJX1
|
four-jointed box kinase 1 |
| chr22_+_25564818 | 0.35 |
ENST00000619906.4
|
GRK3
|
G protein-coupled receptor kinase 3 |
| chr17_-_1486124 | 0.35 |
ENST00000575158.5
|
MYO1C
|
myosin IC |
| chrX_-_49184789 | 0.35 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr1_-_40862354 | 0.35 |
ENST00000372638.4
|
CITED4
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 4 |
| chr11_-_18248662 | 0.35 |
ENST00000256733.9
|
SAA2
|
serum amyloid A2 |
| chr11_+_65919261 | 0.35 |
ENST00000525501.5
|
DRAP1
|
DR1 associated protein 1 |
| chr19_-_14518383 | 0.35 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr11_+_65919687 | 0.34 |
ENST00000532933.1
|
DRAP1
|
DR1 associated protein 1 |
| chr8_+_22999535 | 0.34 |
ENST00000251822.7
|
RHOBTB2
|
Rho related BTB domain containing 2 |
| chr19_-_45423839 | 0.34 |
ENST00000340192.11
|
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr10_-_131981912 | 0.34 |
ENST00000540159.3
|
BNIP3
|
BCL2 interacting protein 3 |
| chr16_+_15643267 | 0.34 |
ENST00000396355.5
|
NDE1
|
nudE neurodevelopment protein 1 |
| chr5_+_60945193 | 0.33 |
ENST00000296597.10
ENST00000511107.1 ENST00000677932.1 ENST00000502658.1 |
NDUFAF2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr2_+_173075435 | 0.33 |
ENST00000539448.5
|
MAP3K20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr11_+_65919331 | 0.33 |
ENST00000376991.6
|
DRAP1
|
DR1 associated protein 1 |
| chrX_-_142205260 | 0.33 |
ENST00000247452.4
|
MAGEC2
|
MAGE family member C2 |
| chr9_-_137070548 | 0.33 |
ENST00000409687.5
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr7_-_44189428 | 0.32 |
ENST00000673284.1
ENST00000403799.8 ENST00000671824.1 |
GCK
|
glucokinase |
| chr10_+_122271292 | 0.32 |
ENST00000260723.6
|
BTBD16
|
BTB domain containing 16 |
| chrX_-_23743201 | 0.32 |
ENST00000492081.1
ENST00000379303.10 ENST00000336430.11 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr19_-_41353904 | 0.32 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr1_-_183635776 | 0.31 |
ENST00000359856.11
|
ARPC5
|
actin related protein 2/3 complex subunit 5 |
| chr16_+_28823212 | 0.31 |
ENST00000570200.5
|
ATXN2L
|
ataxin 2 like |
| chr1_-_71047803 | 0.31 |
ENST00000370932.6
ENST00000628037.2 ENST00000351052.9 ENST00000354608.9 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 |
| chr2_-_88992903 | 0.31 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr9_+_38392694 | 0.31 |
ENST00000377698.4
ENST00000635162.1 |
ALDH1B1
|
aldehyde dehydrogenase 1 family member B1 |
| chr3_-_48595267 | 0.31 |
ENST00000328333.12
ENST00000681320.1 |
COL7A1
|
collagen type VII alpha 1 chain |
| chr1_-_150234668 | 0.31 |
ENST00000616917.4
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr11_-_2903490 | 0.31 |
ENST00000455942.3
ENST00000625099.4 |
SLC22A18AS
|
solute carrier family 22 member 18 antisense |
| chr16_-_68236069 | 0.31 |
ENST00000473183.7
ENST00000565858.5 |
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr8_-_143840940 | 0.31 |
ENST00000442628.7
|
NRBP2
|
nuclear receptor binding protein 2 |
| chr5_+_177303768 | 0.30 |
ENST00000303204.9
ENST00000503216.5 |
PRELID1
|
PRELI domain containing 1 |
| chr16_-_28211908 | 0.30 |
ENST00000566073.1
ENST00000304658.10 |
XPO6
|
exportin 6 |
| chr1_+_28259473 | 0.30 |
ENST00000253063.4
|
SESN2
|
sestrin 2 |
| chr19_-_14529193 | 0.30 |
ENST00000596853.6
ENST00000676515.1 ENST00000678338.1 ENST00000595992.6 ENST00000677848.1 ENST00000677762.1 ENST00000678009.1 ENST00000596075.2 ENST00000601533.6 ENST00000396969.8 ENST00000598692.2 ENST00000678098.1 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr12_+_112938422 | 0.30 |
ENST00000680044.1
ENST00000680966.1 ENST00000548514.2 ENST00000681497.1 ENST00000551007.1 ENST00000228928.12 ENST00000680438.1 ENST00000681147.1 ENST00000679354.1 ENST00000681085.1 ENST00000680161.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3 |
| chr6_+_32637396 | 0.30 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr19_-_48898737 | 0.30 |
ENST00000221399.8
|
TULP2
|
TUB like protein 2 |
| chr20_+_36461747 | 0.30 |
ENST00000340491.8
|
DLGAP4
|
DLG associated protein 4 |
| chr17_-_1485733 | 0.30 |
ENST00000648446.1
|
MYO1C
|
myosin IC |
| chr17_-_44830774 | 0.30 |
ENST00000590758.3
ENST00000591424.5 |
GJC1
|
gap junction protein gamma 1 |
| chr12_+_100267220 | 0.29 |
ENST00000635101.1
|
SCYL2
|
SCY1 like pseudokinase 2 |
| chr19_-_45423501 | 0.28 |
ENST00000591636.5
ENST00000013807.9 ENST00000592023.5 |
ERCC1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr19_-_14517425 | 0.28 |
ENST00000676577.1
ENST00000677204.1 ENST00000598235.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr8_-_61646807 | 0.28 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr16_+_81444799 | 0.28 |
ENST00000537098.8
|
CMIP
|
c-Maf inducing protein |
| chr1_+_184051865 | 0.27 |
ENST00000644815.1
|
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chr1_+_109910892 | 0.27 |
ENST00000369802.7
ENST00000420111.6 |
CSF1
|
colony stimulating factor 1 |
| chr11_+_60924452 | 0.27 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr14_+_20999255 | 0.27 |
ENST00000554422.5
ENST00000298681.5 |
SLC39A2
|
solute carrier family 39 member 2 |
| chr14_-_106875069 | 0.27 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr17_+_4951758 | 0.26 |
ENST00000518175.1
|
ENO3
|
enolase 3 |
| chr1_+_32817442 | 0.26 |
ENST00000373476.5
ENST00000529027.5 |
S100PBP
|
S100P binding protein |
| chr7_-_21945866 | 0.26 |
ENST00000356195.9
ENST00000447180.5 ENST00000373934.4 ENST00000406877.8 ENST00000457951.5 |
CDCA7L
|
cell division cycle associated 7 like |
| chr11_-_76670737 | 0.26 |
ENST00000260061.9
ENST00000404995.5 |
LRRC32
|
leucine rich repeat containing 32 |
| chrX_+_9786420 | 0.26 |
ENST00000380913.8
|
SHROOM2
|
shroom family member 2 |
| chr9_-_68540879 | 0.26 |
ENST00000377311.4
|
TMEM252
|
transmembrane protein 252 |
| chr17_+_7439504 | 0.25 |
ENST00000575331.1
ENST00000293829.9 |
ENSG00000272884.1
FGF11
|
novel transcript fibroblast growth factor 11 |
| chr19_+_535824 | 0.25 |
ENST00000606065.3
|
CDC34
|
cell division cycle 34, ubiqiutin conjugating enzyme |
| chr3_+_141487008 | 0.25 |
ENST00000286364.9
ENST00000452898.2 |
RASA2
|
RAS p21 protein activator 2 |
| chr16_-_46831043 | 0.25 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr19_-_38256339 | 0.25 |
ENST00000591291.5
ENST00000301242.9 |
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr20_-_44187093 | 0.25 |
ENST00000342272.3
|
JPH2
|
junctophilin 2 |
| chrX_+_49171889 | 0.25 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2 |
| chr16_+_70579867 | 0.25 |
ENST00000429149.6
|
IL34
|
interleukin 34 |
| chr17_-_64263221 | 0.24 |
ENST00000258991.7
ENST00000583738.1 ENST00000584379.6 |
TEX2
|
testis expressed 2 |
| chr2_-_74441882 | 0.24 |
ENST00000272430.10
|
RTKN
|
rhotekin |
| chr5_+_177304571 | 0.24 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr1_-_47190013 | 0.24 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr19_-_38256513 | 0.24 |
ENST00000347262.8
ENST00000591585.1 |
PPP1R14A
|
protein phosphatase 1 regulatory inhibitor subunit 14A |
| chr10_+_102394488 | 0.24 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2 |
| chrX_-_15335407 | 0.24 |
ENST00000635543.1
ENST00000542278.6 ENST00000482148.6 ENST00000333590.6 ENST00000637296.1 ENST00000634582.1 ENST00000634640.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis class A |
| chr7_+_101154981 | 0.24 |
ENST00000646560.1
|
AP1S1
|
adaptor related protein complex 1 subunit sigma 1 |
| chr15_+_48331403 | 0.24 |
ENST00000558813.5
ENST00000331200.8 ENST00000558472.5 |
DUT
|
deoxyuridine triphosphatase |
| chr8_+_38820332 | 0.24 |
ENST00000518809.5
ENST00000520611.1 |
TACC1
|
transforming acidic coiled-coil containing protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.4 | 1.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 1.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.4 | 1.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.4 | 1.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 3.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 0.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.3 | 0.8 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.2 | 1.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 1.4 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.9 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 1.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 1.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.4 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 1.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.4 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.6 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.1 | 0.7 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.3 | GO:1902567 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.6 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 1.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 1.2 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.6 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 0.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.7 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.3 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 1.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.6 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 2.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.9 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.7 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 1.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.6 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.4 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.2 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.7 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 1.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.9 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 1.3 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.7 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.8 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:1900222 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 0.7 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.2 | 0.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.2 | 1.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 0.6 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.2 | 1.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 1.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 3.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.7 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 4.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 6.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.3 | 1.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 1.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 1.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 1.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 1.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.7 | GO:0036042 | lysophosphatidic acid binding(GO:0035727) long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.3 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 4 binding(GO:0035662) Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 5.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 2.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 2.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |