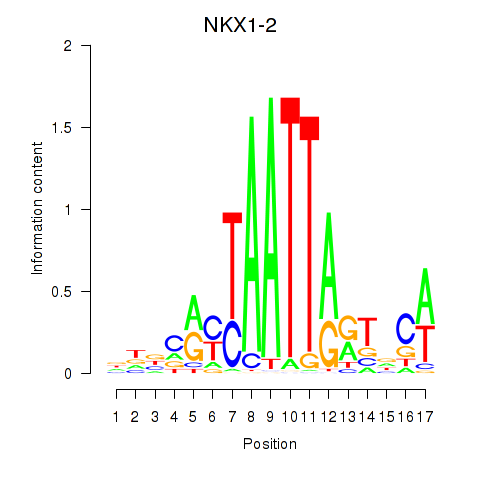
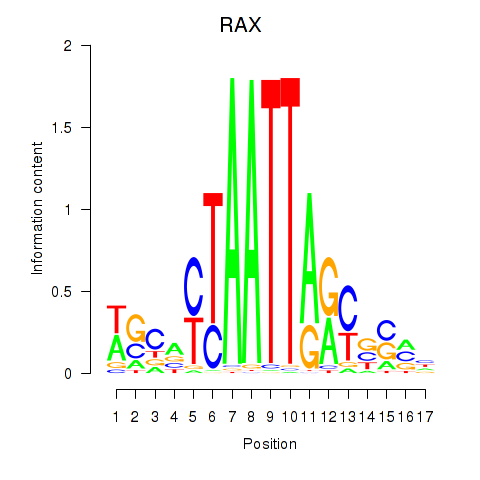
|
chr5_-_35938572
Show fit
|
1.78 |
ENST00000651391.1
ENST00000397366.5
ENST00000513623.5
ENST00000514524.2
ENST00000397367.6
|
CAPSL
|
calcyphosine like
|
|
chr12_+_6904733
Show fit
|
1.12 |
ENST00000007969.12
ENST00000622489.4
ENST00000443597.7
ENST00000323702.9
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr12_+_6904962
Show fit
|
0.98 |
ENST00000415834.5
ENST00000436789.5
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr12_-_25195074
Show fit
|
0.68 |
ENST00000354189.9
ENST00000676236.1
ENST00000545133.5
ENST00000554347.1
ENST00000674567.1
ENST00000395987.8
ENST00000320267.13
ENST00000395990.6
|
CFAP94
|
cilia and flagella associated protein 94
|
|
chr3_-_105869035
Show fit
|
0.57 |
ENST00000447441.6
ENST00000403724.5
ENST00000405772.5
|
CBLB
|
Cbl proto-oncogene B
|
|
chr13_+_35476740
Show fit
|
0.43 |
ENST00000537702.5
|
NBEA
|
neurobeachin
|
|
chr8_-_42768602
Show fit
|
0.29 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr4_+_76435216
Show fit
|
0.29 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr20_-_31390483
Show fit
|
0.28 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119
|
|
chr1_+_160400543
Show fit
|
0.25 |
ENST00000368061.3
|
VANGL2
|
VANGL planar cell polarity protein 2
|
|
chr12_-_9869345
Show fit
|
0.25 |
ENST00000228438.3
|
CLEC2B
|
C-type lectin domain family 2 member B
|
|
chr4_+_94974984
Show fit
|
0.24 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B
|
|
chr7_+_71132123
Show fit
|
0.23 |
ENST00000333538.10
|
GALNT17
|
polypeptide N-acetylgalactosaminyltransferase 17
|
|
chr16_-_28597042
Show fit
|
0.22 |
ENST00000533150.5
ENST00000335715.9
|
SULT1A2
|
sulfotransferase family 1A member 2
|
|
chr11_+_124183219
Show fit
|
0.22 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3
|
|
chr11_+_72189659
Show fit
|
0.21 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha
|
|
chr11_+_72189528
Show fit
|
0.20 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha
|
|
chr9_+_122371014
Show fit
|
0.19 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr5_-_160119389
Show fit
|
0.19 |
ENST00000523662.1
ENST00000456329.7
ENST00000307063.9
|
PWWP2A
|
PWWP domain containing 2A
|
|
chrX_-_152830721
Show fit
|
0.18 |
ENST00000370277.5
|
CETN2
|
centrin 2
|
|
chr2_+_216659600
Show fit
|
0.17 |
ENST00000456764.1
|
IGFBP2
|
insulin like growth factor binding protein 2
|
|
chr9_+_122371036
Show fit
|
0.17 |
ENST00000619306.5
ENST00000426608.6
ENST00000223423.8
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr5_-_20575850
Show fit
|
0.17 |
ENST00000507958.5
|
CDH18
|
cadherin 18
|
|
chr19_+_49513353
Show fit
|
0.16 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter
|
|
chr1_-_92486916
Show fit
|
0.16 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor
|
|
chr17_-_41184895
Show fit
|
0.15 |
ENST00000620667.1
ENST00000398472.2
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr3_-_180679468
Show fit
|
0.15 |
ENST00000651046.1
ENST00000476379.6
|
CCDC39
|
coiled-coil domain containing 39
|
|
chr5_-_83673544
Show fit
|
0.12 |
ENST00000503117.1
ENST00000510978.5
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr3_+_140678041
Show fit
|
0.12 |
ENST00000286349.4
|
TRIM42
|
tripartite motif containing 42
|
|
chrX_+_44844015
Show fit
|
0.12 |
ENST00000339042.6
|
DUSP21
|
dual specificity phosphatase 21
|
|
chr4_-_25863537
Show fit
|
0.12 |
ENST00000502949.5
ENST00000264868.9
ENST00000513691.1
ENST00000514872.1
|
SEL1L3
|
SEL1L family member 3
|
|
chr8_-_100559702
Show fit
|
0.11 |
ENST00000520311.5
ENST00000520552.5
ENST00000521345.1
ENST00000523000.5
ENST00000335659.7
ENST00000358990.3
ENST00000519597.5
|
ANKRD46
|
ankyrin repeat domain 46
|
|
chr11_-_59615673
Show fit
|
0.11 |
ENST00000263847.6
|
OSBP
|
oxysterol binding protein
|
|
chr3_+_186842687
Show fit
|
0.11 |
ENST00000444204.2
ENST00000320741.7
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr8_+_104223344
Show fit
|
0.11 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr5_+_64505981
Show fit
|
0.11 |
ENST00000334025.3
|
RGS7BP
|
regulator of G protein signaling 7 binding protein
|
|
chr14_+_22202561
Show fit
|
0.10 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2
|
|
chr13_+_77741160
Show fit
|
0.10 |
ENST00000314070.9
ENST00000351546.7
|
SLAIN1
|
SLAIN motif family member 1
|
|
chr3_-_115071333
Show fit
|
0.09 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_103562775
Show fit
|
0.09 |
ENST00000230792.7
ENST00000507423.1
|
NUDT12
|
nudix hydrolase 12
|
|
chr11_+_92969651
Show fit
|
0.08 |
ENST00000257068.3
ENST00000528076.1
|
MTNR1B
|
melatonin receptor 1B
|
|
chr15_+_92900338
Show fit
|
0.08 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr16_-_81220370
Show fit
|
0.08 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene)
|
|
chr16_-_55833186
Show fit
|
0.08 |
ENST00000361503.8
ENST00000422046.6
|
CES1
|
carboxylesterase 1
|
|
chr16_-_55833085
Show fit
|
0.08 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1
|
|
chr6_+_26440472
Show fit
|
0.08 |
ENST00000494393.5
ENST00000482451.5
ENST00000471353.5
ENST00000361232.7
ENST00000487627.5
ENST00000496719.1
ENST00000244519.7
ENST00000490254.5
ENST00000487272.1
|
BTN3A3
|
butyrophilin subfamily 3 member A3
|
|
chr4_-_65670478
Show fit
|
0.07 |
ENST00000613740.5
ENST00000622150.4
ENST00000511294.1
|
EPHA5
|
EPH receptor A5
|
|
chr14_-_99480784
Show fit
|
0.07 |
ENST00000329331.7
ENST00000436070.6
|
SETD3
|
SET domain containing 3, actin histidine methyltransferase
|
|
chr9_-_5304713
Show fit
|
0.07 |
ENST00000381627.4
|
RLN2
|
relaxin 2
|
|
chr11_-_7941708
Show fit
|
0.07 |
ENST00000642047.1
|
OR10A3
|
olfactory receptor family 10 subfamily A member 3
|
|
chr4_-_65670339
Show fit
|
0.07 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5
|
|
chr7_-_88226965
Show fit
|
0.07 |
ENST00000490437.5
ENST00000431660.5
|
SRI
|
sorcin
|
|
chr7_-_88226987
Show fit
|
0.07 |
ENST00000394641.7
|
SRI
|
sorcin
|
|
chr7_-_100119840
Show fit
|
0.07 |
ENST00000437822.6
|
TAF6
|
TATA-box binding protein associated factor 6
|
|
chr20_+_45416551
Show fit
|
0.07 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T
|
|
chr9_+_122370523
Show fit
|
0.07 |
ENST00000643810.1
ENST00000540753.6
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr6_+_26402289
Show fit
|
0.06 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1
|
|
chr6_-_31862809
Show fit
|
0.06 |
ENST00000375631.5
|
NEU1
|
neuraminidase 1
|
|
chr1_-_158426237
Show fit
|
0.06 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2
|
|
chr12_-_91180365
Show fit
|
0.06 |
ENST00000547937.5
|
DCN
|
decorin
|
|
chr11_+_19712823
Show fit
|
0.06 |
ENST00000396085.6
ENST00000349880.9
|
NAV2
|
neuron navigator 2
|
|
chr11_-_62754141
Show fit
|
0.06 |
ENST00000527994.1
ENST00000394807.5
ENST00000673933.1
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr15_+_49170200
Show fit
|
0.06 |
ENST00000396509.6
|
GALK2
|
galactokinase 2
|
|
chr8_-_101790934
Show fit
|
0.06 |
ENST00000523645.5
ENST00000520346.1
ENST00000220931.11
ENST00000522448.5
ENST00000522951.5
ENST00000522252.5
ENST00000519098.5
|
NCALD
|
neurocalcin delta
|
|
chr3_-_11643871
Show fit
|
0.06 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4
|
|
chr15_+_49170237
Show fit
|
0.06 |
ENST00000560031.6
ENST00000558145.5
ENST00000544523.5
ENST00000560138.5
|
GALK2
|
galactokinase 2
|
|
chr12_+_26195313
Show fit
|
0.06 |
ENST00000422622.3
|
SSPN
|
sarcospan
|
|
chr19_-_45782388
Show fit
|
0.06 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase
|
|
chr16_+_53099100
Show fit
|
0.05 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr8_+_106726012
Show fit
|
0.05 |
ENST00000449762.6
ENST00000297447.10
|
OXR1
|
oxidation resistance 1
|
|
chr12_-_21910853
Show fit
|
0.05 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9
|
|
chr6_-_166627244
Show fit
|
0.05 |
ENST00000265678.9
|
RPS6KA2
|
ribosomal protein S6 kinase A2
|
|
chr4_-_145180496
Show fit
|
0.05 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4
|
|
chr19_-_9107475
Show fit
|
0.05 |
ENST00000641081.1
|
OR7G2
|
olfactory receptor family 7 subfamily G member 2
|
|
chr9_-_116687205
Show fit
|
0.05 |
ENST00000288520.9
ENST00000358637.4
ENST00000341734.8
|
ASTN2
|
astrotactin 2
|
|
chrX_+_77910656
Show fit
|
0.05 |
ENST00000343533.9
ENST00000341514.11
ENST00000645454.1
ENST00000642651.1
ENST00000644362.1
|
ATP7A
PGK1
|
ATPase copper transporting alpha
phosphoglycerate kinase 1
|
|
chr6_+_54018910
Show fit
|
0.05 |
ENST00000514921.5
ENST00000274897.9
ENST00000370877.6
|
MLIP
|
muscular LMNA interacting protein
|
|
chr1_-_201171545
Show fit
|
0.05 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9
|
|
chr9_+_121567057
Show fit
|
0.05 |
ENST00000394340.7
ENST00000436835.5
ENST00000259371.6
|
DAB2IP
|
DAB2 interacting protein
|
|
chr6_+_26365176
Show fit
|
0.05 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr13_-_110242694
Show fit
|
0.05 |
ENST00000648989.1
ENST00000647797.1
ENST00000648966.1
ENST00000649484.1
ENST00000648695.1
ENST00000650115.1
ENST00000650566.1
|
COL4A1
|
collagen type IV alpha 1 chain
|
|
chr6_+_26365215
Show fit
|
0.04 |
ENST00000527422.5
ENST00000356386.6
ENST00000396948.5
|
BTN3A2
|
butyrophilin subfamily 3 member A2
|
|
chr19_-_45782479
Show fit
|
0.04 |
ENST00000447742.6
ENST00000354227.9
ENST00000291270.9
ENST00000683086.1
ENST00000343373.10
|
DMPK
|
DM1 protein kinase
|
|
chrX_+_89921903
Show fit
|
0.04 |
ENST00000283891.6
ENST00000561129.2
|
TGIF2LX
|
TGFB induced factor homeobox 2 like X-linked
|
|
chr6_+_160121859
Show fit
|
0.04 |
ENST00000324965.8
ENST00000457470.6
|
SLC22A1
|
solute carrier family 22 member 1
|
|
chr15_+_92904447
Show fit
|
0.04 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr1_-_151831768
Show fit
|
0.04 |
ENST00000318247.7
|
RORC
|
RAR related orphan receptor C
|
|
chr10_+_24466487
Show fit
|
0.04 |
ENST00000396446.5
ENST00000396445.5
ENST00000376451.4
|
KIAA1217
|
KIAA1217
|
|
chr7_+_138460238
Show fit
|
0.04 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24
|
|
chr7_+_55365317
Show fit
|
0.04 |
ENST00000254770.3
|
LANCL2
|
LanC like 2
|
|
chr15_+_92900189
Show fit
|
0.04 |
ENST00000626874.2
ENST00000627622.1
ENST00000629346.2
ENST00000628375.2
ENST00000420239.7
ENST00000394196.9
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr14_+_21997531
Show fit
|
0.03 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17
|
|
chr6_+_26402237
Show fit
|
0.03 |
ENST00000476549.6
ENST00000450085.6
ENST00000425234.6
ENST00000427334.5
ENST00000506698.1
ENST00000289361.11
|
BTN3A1
|
butyrophilin subfamily 3 member A1
|
|
chr4_-_119322128
Show fit
|
0.03 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2
|
|
chr7_+_100119607
Show fit
|
0.03 |
ENST00000262932.5
|
CNPY4
|
canopy FGF signaling regulator 4
|
|
chr9_-_5339874
Show fit
|
0.03 |
ENST00000223862.2
|
RLN1
|
relaxin 1
|
|
chr13_-_109786567
Show fit
|
0.03 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2
|
|
chr2_-_238288600
Show fit
|
0.03 |
ENST00000254657.8
|
PER2
|
period circadian regulator 2
|
|
chrX_-_13817027
Show fit
|
0.03 |
ENST00000493677.5
ENST00000355135.6
ENST00000316715.9
|
GPM6B
|
glycoprotein M6B
|
|
chr19_+_35748549
Show fit
|
0.03 |
ENST00000301159.14
|
LIN37
|
lin-37 DREAM MuvB core complex component
|
|
chr8_+_91249307
Show fit
|
0.03 |
ENST00000309536.6
ENST00000276609.8
|
SLC26A7
|
solute carrier family 26 member 7
|
|
chr19_+_49487510
Show fit
|
0.03 |
ENST00000679106.1
ENST00000621674.4
ENST00000391857.9
ENST00000678510.1
ENST00000467825.2
|
RPL13A
|
ribosomal protein L13a
|
|
chr15_+_94355956
Show fit
|
0.03 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2
|
|
chr7_+_100049765
Show fit
|
0.03 |
ENST00000456748.6
ENST00000292450.9
ENST00000438937.1
ENST00000543588.2
|
ZSCAN21
|
zinc finger and SCAN domain containing 21
|
|
chr16_-_20691256
Show fit
|
0.03 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1
|
|
chr6_+_125153649
Show fit
|
0.02 |
ENST00000304877.17
ENST00000368402.9
ENST00000368388.6
ENST00000534000.6
|
TPD52L1
|
TPD52 like 1
|
|
chr14_-_99480831
Show fit
|
0.02 |
ENST00000331768.10
ENST00000630307.2
|
SETD3
|
SET domain containing 3, actin histidine methyltransferase
|
|
chr17_-_10469558
Show fit
|
0.02 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4
|
|
chr7_-_87220567
Show fit
|
0.02 |
ENST00000433078.5
|
TMEM243
|
transmembrane protein 243
|
|
chr6_+_29550407
Show fit
|
0.02 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene
|
|
chr19_-_3985451
Show fit
|
0.02 |
ENST00000309311.7
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr6_-_111606260
Show fit
|
0.02 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr12_+_8157034
Show fit
|
0.02 |
ENST00000396570.7
|
ZNF705A
|
zinc finger protein 705A
|
|
chr1_+_226063466
Show fit
|
0.02 |
ENST00000666609.1
ENST00000661429.1
|
H3-3A
|
H3.3 histone A
|
|
chr8_+_104223320
Show fit
|
0.02 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr19_+_49766962
Show fit
|
0.02 |
ENST00000354293.10
ENST00000359032.9
|
AP2A1
|
adaptor related protein complex 2 subunit alpha 1
|
|
chr6_+_154039102
Show fit
|
0.02 |
ENST00000360422.8
ENST00000330432.12
|
OPRM1
|
opioid receptor mu 1
|
|
chr19_+_54137740
Show fit
|
0.02 |
ENST00000644245.1
ENST00000646002.1
ENST00000221232.11
ENST00000440571.6
ENST00000617930.2
|
CNOT3
|
CCR4-NOT transcription complex subunit 3
|
|
chr6_+_30720335
Show fit
|
0.02 |
ENST00000327892.13
|
TUBB
|
tubulin beta class I
|
|
chrX_+_77899440
Show fit
|
0.02 |
ENST00000373335.4
ENST00000475465.1
ENST00000650309.2
ENST00000647835.1
|
COX7B
|
cytochrome c oxidase subunit 7B
|
|
chrX_+_136536099
Show fit
|
0.02 |
ENST00000440515.5
ENST00000456412.1
|
VGLL1
|
vestigial like family member 1
|
|
chr3_-_151316795
Show fit
|
0.02 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87
|
|
chr2_-_86194972
Show fit
|
0.02 |
ENST00000254636.9
|
IMMT
|
inner membrane mitochondrial protein
|
|
chr2_+_89884740
Show fit
|
0.02 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional)
|
|
chr12_-_55296569
Show fit
|
0.02 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6
|
|
chr1_+_42682388
Show fit
|
0.02 |
ENST00000321358.12
ENST00000332220.10
|
YBX1
|
Y-box binding protein 1
|
|
chr2_-_89297785
Show fit
|
0.02 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional)
|
|
chr17_-_4967790
Show fit
|
0.02 |
ENST00000575142.5
ENST00000206020.8
|
SPAG7
|
sperm associated antigen 7
|
|
chr17_-_5035418
Show fit
|
0.02 |
ENST00000254853.10
ENST00000424747.1
|
SLC52A1
|
solute carrier family 52 member 1
|
|
chr1_+_100352451
Show fit
|
0.01 |
ENST00000361544.11
ENST00000370124.8
ENST00000336454.5
|
CDC14A
|
cell division cycle 14A
|
|
chr7_-_100119323
Show fit
|
0.01 |
ENST00000523306.5
ENST00000344095.8
ENST00000417349.5
ENST00000493322.5
ENST00000520135.5
ENST00000460673.2
ENST00000453269.7
ENST00000452041.5
ENST00000452438.6
ENST00000451699.5
|
TAF6
|
TATA-box binding protein associated factor 6
|
|
chr16_+_3283443
Show fit
|
0.01 |
ENST00000572748.1
ENST00000219069.6
ENST00000573578.1
ENST00000574253.1
|
ZNF263
|
zinc finger protein 263
|
|
chr1_-_152414256
Show fit
|
0.01 |
ENST00000271835.3
|
CRNN
|
cornulin
|
|
chr8_+_106726115
Show fit
|
0.01 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1
|
|
chr10_+_18260715
Show fit
|
0.01 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr4_-_73620629
Show fit
|
0.01 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6
|
|
chr1_+_27934980
Show fit
|
0.01 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B
|
|
chr12_+_16347637
Show fit
|
0.01 |
ENST00000543076.5
ENST00000396210.8
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr17_-_41060109
Show fit
|
0.01 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3
|
|
chr17_+_7407838
Show fit
|
0.01 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2
|
|
chr9_-_134068012
Show fit
|
0.01 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3
|
|
chr9_+_34329545
Show fit
|
0.01 |
ENST00000379158.7
|
NUDT2
|
nudix hydrolase 2
|
|
chr19_+_10111689
Show fit
|
0.01 |
ENST00000321826.5
|
P2RY11
|
purinergic receptor P2Y11
|
|
chr6_-_111605859
Show fit
|
0.00 |
ENST00000651359.1
ENST00000650859.1
ENST00000359831.8
ENST00000368761.11
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr3_-_139006268
Show fit
|
0.00 |
ENST00000383163.4
|
PRR23A
|
proline rich 23A
|
|
chr5_+_67004618
Show fit
|
0.00 |
ENST00000261569.11
ENST00000436277.5
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr12_-_118359105
Show fit
|
0.00 |
ENST00000541186.5
ENST00000539872.5
|
TAOK3
|
TAO kinase 3
|
|
chr1_+_45688165
Show fit
|
0.00 |
ENST00000372025.5
|
TMEM69
|
transmembrane protein 69
|
|
chr2_-_74391837
Show fit
|
0.00 |
ENST00000417090.1
ENST00000409868.5
ENST00000680606.1
|
DCTN1
|
dynactin subunit 1
|
|
chr3_-_191282383
Show fit
|
0.00 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|