Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for NR1D1
Z-value: 0.38
Transcription factors associated with NR1D1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1D1
|
ENSG00000126368.6 | NR1D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1D1 | hg38_v1_chr17_-_40100569_40100597 | -0.22 | 2.3e-01 | Click! |
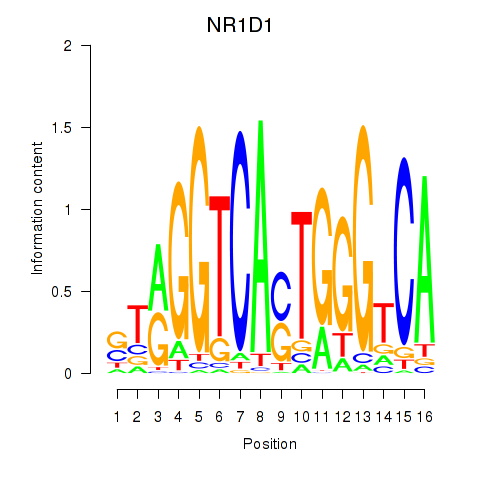
Activity profile of NR1D1 motif
Sorted Z-values of NR1D1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1D1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_20589044 | 1.81 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr1_-_25906457 | 1.29 |
ENST00000426559.6
|
STMN1
|
stathmin 1 |
| chr11_-_125496122 | 1.25 |
ENST00000527534.1
ENST00000278919.8 ENST00000366139.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 |
| chr19_-_42423100 | 0.88 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type |
| chr2_+_126898857 | 0.85 |
ENST00000643416.1
|
TEX51
|
testis expressed 51 |
| chr1_-_6497096 | 0.71 |
ENST00000537245.6
|
PLEKHG5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr9_-_33167296 | 0.69 |
ENST00000379731.5
ENST00000535206.5 |
B4GALT1
|
beta-1,4-galactosyltransferase 1 |
| chr2_+_207711534 | 0.64 |
ENST00000392209.7
|
CCNYL1
|
cyclin Y like 1 |
| chr1_-_111449209 | 0.61 |
ENST00000235090.10
|
WDR77
|
WD repeat domain 77 |
| chr14_-_91244669 | 0.60 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr14_+_24070837 | 0.60 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr14_-_94129577 | 0.57 |
ENST00000238609.4
|
IFI27L2
|
interferon alpha inducible protein 27 like 2 |
| chr1_-_25906411 | 0.54 |
ENST00000455785.7
|
STMN1
|
stathmin 1 |
| chr2_+_126898908 | 0.53 |
ENST00000450035.5
|
TEX51
|
testis expressed 51 |
| chr2_+_126898876 | 0.52 |
ENST00000568484.6
ENST00000636457.1 |
TEX51
|
testis expressed 51 |
| chr4_+_168921555 | 0.52 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_43313473 | 0.51 |
ENST00000433297.2
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr5_-_43313403 | 0.50 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr3_-_182985926 | 0.49 |
ENST00000487822.5
ENST00000460412.6 ENST00000469954.5 |
DCUN1D1
|
defective in cullin neddylation 1 domain containing 1 |
| chr1_-_25905989 | 0.46 |
ENST00000399728.5
|
STMN1
|
stathmin 1 |
| chr22_+_22922594 | 0.45 |
ENST00000390331.3
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr2_+_207711631 | 0.45 |
ENST00000295414.8
ENST00000420822.1 ENST00000339882.9 |
CCNYL1
|
cyclin Y like 1 |
| chr16_+_71358713 | 0.42 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr19_-_43198079 | 0.41 |
ENST00000597374.5
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr22_-_37484505 | 0.38 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_-_61863327 | 0.38 |
ENST00000584322.2
ENST00000682369.1 ENST00000683039.1 ENST00000683381.1 |
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr1_-_25906931 | 0.38 |
ENST00000357865.6
|
STMN1
|
stathmin 1 |
| chr3_-_185821092 | 0.37 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chrX_+_54920796 | 0.37 |
ENST00000442098.5
ENST00000430420.5 ENST00000453081.5 ENST00000319167.12 ENST00000622017.4 ENST00000375022.8 ENST00000399736.5 ENST00000440072.5 ENST00000173898.12 ENST00000431115.5 ENST00000440759.5 ENST00000375041.6 |
TRO
|
trophinin |
| chr1_+_43933277 | 0.36 |
ENST00000414809.7
|
ARTN
|
artemin |
| chr1_+_2467437 | 0.35 |
ENST00000449969.5
|
PLCH2
|
phospholipase C eta 2 |
| chr19_-_14518383 | 0.35 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr6_+_31575557 | 0.34 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor |
| chr1_-_11805924 | 0.34 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr10_-_77140757 | 0.32 |
ENST00000637862.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_-_11805977 | 0.31 |
ENST00000376486.3
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr19_-_43204223 | 0.29 |
ENST00000599746.5
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr11_+_60924452 | 0.29 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr3_+_155083523 | 0.29 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr9_-_133376085 | 0.28 |
ENST00000485435.6
|
SURF4
|
surfeit 4 |
| chr8_+_11704151 | 0.28 |
ENST00000526716.5
ENST00000532059.6 ENST00000335135.8 ENST00000622443.3 |
GATA4
|
GATA binding protein 4 |
| chr19_-_49361475 | 0.27 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2 |
| chr2_+_90114838 | 0.26 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr3_+_172040554 | 0.26 |
ENST00000336824.8
ENST00000423424.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr8_+_104223320 | 0.26 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr16_+_3654683 | 0.25 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr1_-_11805949 | 0.22 |
ENST00000376590.9
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr1_-_11805294 | 0.22 |
ENST00000413656.5
ENST00000376592.6 ENST00000376585.6 |
MTHFR
|
methylenetetrahydrofolate reductase |
| chr1_+_161749762 | 0.22 |
ENST00000367943.5
|
DUSP12
|
dual specificity phosphatase 12 |
| chr10_+_11005301 | 0.21 |
ENST00000416382.6
ENST00000631460.1 ENST00000631816.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr18_+_33578213 | 0.21 |
ENST00000681521.1
ENST00000269197.12 |
ASXL3
|
ASXL transcriptional regulator 3 |
| chr7_+_45000184 | 0.20 |
ENST00000544363.5
ENST00000541586.5 ENST00000258781.11 |
CCM2
|
CCM2 scaffold protein |
| chr22_+_37019735 | 0.19 |
ENST00000429360.6
ENST00000341116.7 ENST00000404393.5 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr17_-_18644418 | 0.19 |
ENST00000575220.5
ENST00000405044.6 ENST00000573652.1 |
TBC1D28
|
TBC1 domain family member 28 |
| chr22_+_26621952 | 0.17 |
ENST00000354760.4
|
CRYBA4
|
crystallin beta A4 |
| chr19_+_21142058 | 0.17 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chr1_+_168280872 | 0.17 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19 |
| chr19_+_21082140 | 0.16 |
ENST00000616183.1
ENST00000596053.5 |
ZNF714
|
zinc finger protein 714 |
| chr10_+_88586762 | 0.15 |
ENST00000371939.7
|
LIPJ
|
lipase family member J |
| chr3_-_51968387 | 0.15 |
ENST00000490063.5
ENST00000468324.5 ENST00000497653.5 ENST00000484633.5 |
PCBP4
|
poly(rC) binding protein 4 |
| chr4_+_87608529 | 0.14 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr1_-_226186673 | 0.14 |
ENST00000366812.6
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr17_+_41255384 | 0.14 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr3_-_52826834 | 0.14 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr17_+_82235769 | 0.13 |
ENST00000619321.2
|
SLC16A3
|
solute carrier family 16 member 3 |
| chr15_-_80989792 | 0.13 |
ENST00000261758.6
ENST00000561312.5 |
MESD
|
mesoderm development LRP chaperone |
| chr9_+_35490103 | 0.13 |
ENST00000361226.8
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr4_-_21697755 | 0.13 |
ENST00000382148.7
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr17_-_78132407 | 0.13 |
ENST00000322914.7
|
TMC6
|
transmembrane channel like 6 |
| chr19_+_21020675 | 0.13 |
ENST00000595401.1
|
ZNF430
|
zinc finger protein 430 |
| chr17_+_42854078 | 0.13 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase copper containing 3 |
| chr1_+_203795614 | 0.13 |
ENST00000367210.3
ENST00000432282.5 ENST00000453771.5 ENST00000367214.5 ENST00000639812.1 ENST00000367212.7 ENST00000332127.8 ENST00000550078.2 |
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A zinc finger BED-type containing 6 |
| chr6_+_167111789 | 0.12 |
ENST00000400926.5
|
CCR6
|
C-C motif chemokine receptor 6 |
| chr17_+_41237998 | 0.12 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr10_+_133453936 | 0.12 |
ENST00000640237.1
|
SCART1
|
scavenger receptor family member expressed on T cells 1 |
| chr1_+_109712272 | 0.12 |
ENST00000369812.6
|
GSTM5
|
glutathione S-transferase mu 5 |
| chr14_+_100323332 | 0.12 |
ENST00000361529.5
ENST00000557052.1 |
SLC25A47
|
solute carrier family 25 member 47 |
| chr12_-_110445540 | 0.12 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr1_+_197413827 | 0.11 |
ENST00000367397.1
ENST00000681519.1 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr15_+_65611336 | 0.11 |
ENST00000537259.5
|
SLC24A1
|
solute carrier family 24 member 1 |
| chr1_-_12618198 | 0.11 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr9_+_136327526 | 0.11 |
ENST00000440944.6
|
GPSM1
|
G protein signaling modulator 1 |
| chr3_-_48685835 | 0.10 |
ENST00000439518.5
ENST00000416649.6 ENST00000294129.7 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr9_-_133376111 | 0.09 |
ENST00000545297.5
ENST00000613129.4 ENST00000618229.4 |
SURF4
|
surfeit 4 |
| chr7_-_129054869 | 0.09 |
ENST00000471166.1
|
TNPO3
|
transportin 3 |
| chr17_+_45844875 | 0.08 |
ENST00000329196.7
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr1_+_109712227 | 0.08 |
ENST00000256593.8
|
GSTM5
|
glutathione S-transferase mu 5 |
| chr1_-_168543990 | 0.08 |
ENST00000367819.3
|
XCL2
|
X-C motif chemokine ligand 2 |
| chr17_+_6070361 | 0.07 |
ENST00000317744.10
|
WSCD1
|
WSC domain containing 1 |
| chr2_+_88885397 | 0.07 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr10_-_102120318 | 0.07 |
ENST00000673968.1
|
LDB1
|
LIM domain binding 1 |
| chr6_+_75749272 | 0.07 |
ENST00000653423.1
|
MYO6
|
myosin VI |
| chr20_+_48921775 | 0.06 |
ENST00000681021.1
ENST00000679436.1 |
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr7_+_114922561 | 0.06 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr5_-_63962438 | 0.05 |
ENST00000323865.5
ENST00000506598.1 |
HTR1A
|
5-hydroxytryptamine receptor 1A |
| chr18_-_46757012 | 0.05 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr17_-_76532046 | 0.05 |
ENST00000590175.5
|
CYGB
|
cytoglobin |
| chr7_+_98106852 | 0.05 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr19_-_18884219 | 0.04 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr7_-_144410227 | 0.04 |
ENST00000467773.1
ENST00000483238.5 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr1_+_159826860 | 0.04 |
ENST00000289707.10
|
SLAMF8
|
SLAM family member 8 |
| chr6_+_75749231 | 0.04 |
ENST00000664209.1
ENST00000369975.6 ENST00000627432.3 ENST00000369985.9 ENST00000369977.8 ENST00000664640.1 ENST00000662603.1 |
MYO6
|
myosin VI |
| chr6_+_75749192 | 0.03 |
ENST00000369981.7
|
MYO6
|
myosin VI |
| chr10_-_102120246 | 0.03 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chr12_-_7018465 | 0.03 |
ENST00000261407.9
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr6_-_169250825 | 0.03 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr11_+_66857056 | 0.03 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr17_-_58280928 | 0.02 |
ENST00000225275.4
|
MPO
|
myeloperoxidase |
| chr16_+_77191173 | 0.02 |
ENST00000248248.8
ENST00000439557.6 ENST00000545553.1 |
MON1B
|
MON1 homolog B, secretory trafficking associated |
| chr9_-_37592564 | 0.02 |
ENST00000544379.1
ENST00000321301.7 ENST00000377773.9 ENST00000401811.3 |
TOMM5
|
translocase of outer mitochondrial membrane 5 |
| chr13_+_27270814 | 0.02 |
ENST00000241463.5
|
RASL11A
|
RAS like family 11 member A |
| chr20_+_48921701 | 0.02 |
ENST00000371917.5
|
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr1_-_10947870 | 0.00 |
ENST00000468348.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.7 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 2.7 | GO:1905098 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 1.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.2 | 0.6 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 1.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.4 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.3 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.1 | 0.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.0 | 0.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 1.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.7 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 0.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 1.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.7 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.6 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |