Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
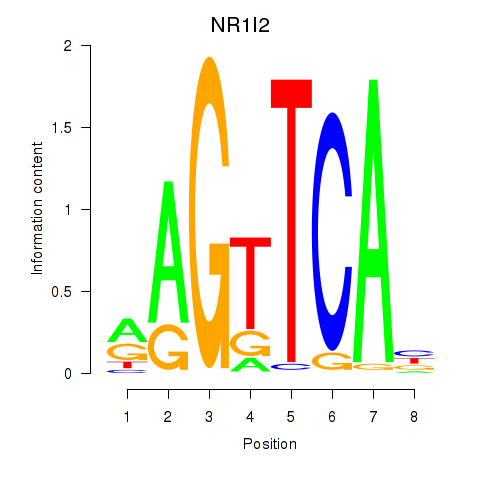
Results for NR1I2
Z-value: 0.66
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.20 | NR1I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I2 | hg38_v1_chr3_+_119782094_119782114 | -0.12 | 5.2e-01 | Click! |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_50968966 | 2.63 |
ENST00000376851.7
|
KLK6
|
kallikrein related peptidase 6 |
| chr1_-_205449924 | 2.47 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr2_-_31217511 | 1.39 |
ENST00000403897.4
|
CAPN14
|
calpain 14 |
| chr20_+_3796288 | 1.35 |
ENST00000439880.6
ENST00000245960.10 |
CDC25B
|
cell division cycle 25B |
| chr19_+_2096960 | 1.33 |
ENST00000588003.5
|
IZUMO4
|
IZUMO family member 4 |
| chr10_+_93073873 | 1.26 |
ENST00000224356.5
|
CYP26A1
|
cytochrome P450 family 26 subfamily A member 1 |
| chr4_-_158173004 | 1.14 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr20_-_54173976 | 1.14 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr2_+_30231524 | 1.05 |
ENST00000395323.9
ENST00000406087.5 ENST00000404397.5 |
LBH
|
LBH regulator of WNT signaling pathway |
| chr2_-_191847068 | 1.03 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2 |
| chr15_+_39581068 | 0.99 |
ENST00000397591.2
ENST00000260356.6 |
THBS1
|
thrombospondin 1 |
| chr4_-_158173042 | 0.97 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr19_+_34994778 | 0.94 |
ENST00000599564.5
|
GRAMD1A
|
GRAM domain containing 1A |
| chr1_+_150549734 | 0.94 |
ENST00000674043.1
ENST00000674058.1 |
ADAMTSL4
|
ADAMTS like 4 |
| chrX_+_136197020 | 0.91 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr10_-_119536533 | 0.87 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr19_+_2096873 | 0.87 |
ENST00000395307.6
ENST00000395301.8 ENST00000620263.4 |
IZUMO4
|
IZUMO family member 4 |
| chr16_+_46884675 | 0.85 |
ENST00000562132.5
ENST00000440783.2 |
GPT2
|
glutamic--pyruvic transaminase 2 |
| chrX_+_136197039 | 0.85 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_15756659 | 0.84 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_-_121800558 | 0.84 |
ENST00000546227.5
|
RHOF
|
ras homolog family member F, filopodia associated |
| chrX_+_136196750 | 0.83 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_148679734 | 0.82 |
ENST00000606877.2
ENST00000593495.3 |
NBPF14
NOTCH2NLB
|
NBPF member 14 notch 2 N-terminal like B |
| chr7_+_134891400 | 0.82 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr1_-_153549120 | 0.79 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_+_120723939 | 0.76 |
ENST00000624419.2
ENST00000652763.1 ENST00000620612.5 |
NOTCH2NLR
NBPF26
|
notch 2 N-terminal like R (pseudogene) NBPF member 26 |
| chr21_-_5973383 | 0.74 |
ENST00000464664.3
|
ENSG00000274559.3
|
novel histone H2B family protein |
| chr19_+_2096937 | 0.72 |
ENST00000395296.5
|
IZUMO4
|
IZUMO family member 4 |
| chrX_-_135098695 | 0.72 |
ENST00000433425.4
|
SMIM10L2B
|
small integral membrane protein 10 like 2B |
| chr3_-_139539577 | 0.70 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr9_-_120876356 | 0.69 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr11_+_130448633 | 0.69 |
ENST00000299164.4
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif 15 |
| chr14_-_58427134 | 0.69 |
ENST00000555930.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr3_+_155083889 | 0.68 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr11_+_121102666 | 0.67 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr3_+_155083523 | 0.65 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr8_-_61646807 | 0.65 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr1_-_146229000 | 0.62 |
ENST00000612520.2
ENST00000579793.6 ENST00000362074.7 |
NBPF10
NOTCH2NLA
|
NBPF member 10 notch 2 N-terminal like A |
| chr2_-_55296361 | 0.62 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_-_52192007 | 0.61 |
ENST00000394815.3
|
KRT80
|
keratin 80 |
| chr12_-_95116967 | 0.61 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_-_52191981 | 0.60 |
ENST00000313234.9
|
KRT80
|
keratin 80 |
| chr1_+_15152522 | 0.60 |
ENST00000428417.5
|
TMEM51
|
transmembrane protein 51 |
| chrX_+_65667645 | 0.58 |
ENST00000360270.7
|
MSN
|
moesin |
| chr20_+_44715360 | 0.58 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr1_-_153549238 | 0.57 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr8_-_81112055 | 0.57 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr7_-_42152396 | 0.57 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr1_+_15152558 | 0.56 |
ENST00000376014.7
ENST00000451326.6 |
TMEM51
|
transmembrane protein 51 |
| chr4_-_110198650 | 0.56 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_110198579 | 0.55 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr11_+_62880863 | 0.55 |
ENST00000680297.1
|
SLC3A2
|
solute carrier family 3 member 2 |
| chr11_+_62880885 | 0.55 |
ENST00000541372.1
ENST00000539458.1 ENST00000338663.12 ENST00000681232.1 ENST00000681657.1 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr16_-_2720217 | 0.54 |
ENST00000302641.8
|
PRSS27
|
serine protease 27 |
| chr18_+_31447732 | 0.54 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr21_+_42403856 | 0.54 |
ENST00000291535.11
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr2_+_236569817 | 0.53 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3 |
| chr11_+_118607579 | 0.51 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr4_+_147732070 | 0.48 |
ENST00000336498.8
|
ARHGAP10
|
Rho GTPase activating protein 10 |
| chr19_-_43204223 | 0.47 |
ENST00000599746.5
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr1_+_74235377 | 0.47 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr8_-_109974688 | 0.47 |
ENST00000297404.1
|
KCNV1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr7_-_42152444 | 0.47 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3 |
| chr12_-_95790755 | 0.47 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr11_+_62881686 | 0.47 |
ENST00000536981.6
ENST00000539891.6 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr13_+_32031300 | 0.47 |
ENST00000642040.1
|
FRY
|
FRY microtubule binding protein |
| chr5_+_33936386 | 0.46 |
ENST00000330120.5
|
RXFP3
|
relaxin family peptide receptor 3 |
| chr6_+_37929959 | 0.46 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr15_-_89211803 | 0.46 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr19_-_6393205 | 0.45 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr3_+_38165484 | 0.45 |
ENST00000446845.5
ENST00000311806.8 |
OXSR1
|
oxidative stress responsive kinase 1 |
| chr3_+_12288838 | 0.45 |
ENST00000455517.6
ENST00000681982.1 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr16_+_67173935 | 0.44 |
ENST00000566871.5
|
NOL3
|
nucleolar protein 3 |
| chr1_+_196819731 | 0.44 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr5_-_1801284 | 0.43 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr5_+_66828762 | 0.43 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_6393131 | 0.43 |
ENST00000394456.10
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr10_-_24721866 | 0.43 |
ENST00000416305.1
ENST00000320481.10 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr6_+_106098933 | 0.43 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr11_+_118607598 | 0.43 |
ENST00000600882.6
ENST00000356063.9 |
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr20_-_13990609 | 0.42 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr1_+_218346235 | 0.42 |
ENST00000366929.4
|
TGFB2
|
transforming growth factor beta 2 |
| chr1_-_231421146 | 0.42 |
ENST00000667629.1
ENST00000670301.1 ENST00000658954.1 |
EGLN1
|
egl-9 family hypoxia inducible factor 1 |
| chr19_-_45153852 | 0.42 |
ENST00000589776.1
|
NKPD1
|
NTPase KAP family P-loop domain containing 1 |
| chr4_-_119322128 | 0.41 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr1_-_149842736 | 0.41 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr7_-_76618300 | 0.41 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chrX_+_50067576 | 0.41 |
ENST00000376108.7
|
CLCN5
|
chloride voltage-gated channel 5 |
| chr12_-_7936177 | 0.41 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr14_-_58427158 | 0.39 |
ENST00000555097.1
ENST00000556367.6 ENST00000555404.5 |
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr7_-_94656197 | 0.39 |
ENST00000643903.1
ENST00000644122.1 ENST00000447873.6 ENST00000644816.1 ENST00000644375.1 |
SGCE
|
sarcoglycan epsilon |
| chr11_-_111912871 | 0.39 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr7_-_94656160 | 0.38 |
ENST00000644609.1
ENST00000643272.1 ENST00000646137.1 ENST00000646098.1 ENST00000643193.1 ENST00000437425.7 ENST00000644551.1 ENST00000415788.3 |
SGCE
|
sarcoglycan epsilon |
| chr3_+_25428233 | 0.38 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chr17_+_40341388 | 0.37 |
ENST00000394086.7
|
RARA
|
retinoic acid receptor alpha |
| chr2_+_233718734 | 0.37 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4 |
| chr12_-_89352487 | 0.37 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr5_-_128339191 | 0.37 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chr3_+_12289061 | 0.37 |
ENST00000652522.1
ENST00000652431.1 ENST00000652098.1 ENST00000651735.1 ENST00000397026.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr1_-_29181809 | 0.36 |
ENST00000466448.4
ENST00000373795.7 |
SRSF4
|
serine and arginine rich splicing factor 4 |
| chr7_+_112450451 | 0.36 |
ENST00000429071.5
ENST00000403825.8 ENST00000675268.1 |
IFRD1
ENSG00000288634.1
|
interferon related developmental regulator 1 novel protein |
| chr10_-_101588126 | 0.36 |
ENST00000339310.7
ENST00000299206.8 ENST00000413344.5 ENST00000429502.1 ENST00000430045.1 ENST00000370172.5 ENST00000370162.8 ENST00000628479.2 |
POLL
|
DNA polymerase lambda |
| chr18_+_63775369 | 0.36 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr16_+_67173971 | 0.36 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 |
| chr6_+_42929127 | 0.36 |
ENST00000394142.7
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr18_+_63775395 | 0.36 |
ENST00000398019.7
|
SERPINB7
|
serpin family B member 7 |
| chr3_+_8501846 | 0.35 |
ENST00000454244.4
|
LMCD1
|
LIM and cysteine rich domains 1 |
| chr1_+_149390612 | 0.35 |
ENST00000650865.1
ENST00000652191.1 ENST00000621744.4 |
NOTCH2NLC
ENSG00000286185.1
|
notch 2 N-terminal like C novel protein, identical to neuroblastoma breakpoint family, member 19 NBPF19 |
| chr5_-_181242236 | 0.35 |
ENST00000507756.5
|
RACK1
|
receptor for activated C kinase 1 |
| chr1_+_115029823 | 0.35 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr13_-_113864062 | 0.34 |
ENST00000327773.7
|
GAS6
|
growth arrest specific 6 |
| chr10_-_75235917 | 0.34 |
ENST00000469299.1
ENST00000372538.8 |
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr19_+_49043837 | 0.34 |
ENST00000301408.7
|
CGB5
|
chorionic gonadotropin subunit beta 5 |
| chr7_-_94655993 | 0.34 |
ENST00000647110.1
ENST00000647048.1 ENST00000643020.1 ENST00000644682.1 ENST00000646119.1 ENST00000646265.1 ENST00000645445.1 ENST00000647334.1 ENST00000645262.1 ENST00000428696.7 ENST00000647096.1 ENST00000642394.1 ENST00000645725.1 ENST00000647351.1 ENST00000646943.1 ENST00000648936.2 ENST00000642707.1 ENST00000645109.1 ENST00000646489.1 ENST00000642933.1 ENST00000643128.1 ENST00000445866.7 ENST00000645101.1 ENST00000644116.1 ENST00000642441.1 ENST00000646879.1 ENST00000647018.1 |
SGCE
|
sarcoglycan epsilon |
| chr3_-_142029108 | 0.34 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr14_-_58427114 | 0.34 |
ENST00000556007.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr21_+_42403874 | 0.33 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr3_+_111911604 | 0.33 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr16_+_83953232 | 0.33 |
ENST00000565123.5
ENST00000393306.6 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr6_+_26272923 | 0.33 |
ENST00000377733.4
|
H2BC10
|
H2B clustered histone 10 |
| chr18_+_23992773 | 0.33 |
ENST00000304621.10
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr22_+_20774092 | 0.32 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr2_-_208190001 | 0.32 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr22_-_38302990 | 0.32 |
ENST00000612795.2
ENST00000451964.5 |
CSNK1E
|
casein kinase 1 epsilon |
| chr2_+_233712905 | 0.31 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr13_+_32031706 | 0.31 |
ENST00000542859.6
|
FRY
|
FRY microtubule binding protein |
| chr6_+_31547560 | 0.31 |
ENST00000376148.9
ENST00000376145.8 |
NFKBIL1
|
NFKB inhibitor like 1 |
| chr2_+_223051814 | 0.30 |
ENST00000281830.3
|
KCNE4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr1_+_43358998 | 0.30 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr1_+_196943738 | 0.30 |
ENST00000367415.8
ENST00000367421.5 ENST00000649283.1 ENST00000476712.6 ENST00000496448.6 ENST00000473386.1 ENST00000649960.1 |
CFHR2
|
complement factor H related 2 |
| chr14_-_73027077 | 0.30 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr22_-_38097772 | 0.29 |
ENST00000332536.10
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr1_+_43358968 | 0.29 |
ENST00000310955.11
|
CDC20
|
cell division cycle 20 |
| chr11_-_111923722 | 0.29 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr14_+_76826372 | 0.29 |
ENST00000393774.7
ENST00000555189.1 |
LRRC74A
|
leucine rich repeat containing 74A |
| chr19_-_51723968 | 0.29 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr1_+_11736068 | 0.29 |
ENST00000376637.7
|
AGTRAP
|
angiotensin II receptor associated protein |
| chr1_+_11736120 | 0.28 |
ENST00000400895.6
ENST00000314340.10 ENST00000376629.8 ENST00000376627.6 ENST00000452018.6 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor associated protein |
| chr16_-_1884231 | 0.28 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chr15_-_64381431 | 0.28 |
ENST00000558008.3
ENST00000300035.9 ENST00000559519.5 ENST00000380258.6 |
PCLAF
|
PCNA clamp associated factor |
| chr13_-_32538683 | 0.28 |
ENST00000674456.1
ENST00000504114.5 |
N4BP2L2
|
NEDD4 binding protein 2 like 2 |
| chr1_-_39883434 | 0.28 |
ENST00000541099.5
ENST00000441669.6 ENST00000316891.10 ENST00000537440.5 ENST00000372818.5 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chr19_+_42313298 | 0.28 |
ENST00000301204.8
ENST00000673205.1 |
TMEM145
|
transmembrane protein 145 |
| chr14_-_73027117 | 0.28 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr7_-_32892015 | 0.28 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr17_-_48604959 | 0.28 |
ENST00000225648.4
ENST00000484302.3 |
HOXB6
|
homeobox B6 |
| chr8_+_144095054 | 0.27 |
ENST00000318911.5
|
CYC1
|
cytochrome c1 |
| chrX_+_100584928 | 0.27 |
ENST00000373031.5
|
TNMD
|
tenomodulin |
| chr4_+_5711154 | 0.27 |
ENST00000264956.11
ENST00000509451.1 |
EVC
|
EvC ciliary complex subunit 1 |
| chr1_+_27934980 | 0.27 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr1_-_197067234 | 0.27 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr14_-_102509798 | 0.27 |
ENST00000560748.5
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr16_+_71358713 | 0.26 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr1_-_248645278 | 0.26 |
ENST00000641268.1
|
OR2T35
|
olfactory receptor family 2 subfamily T member 35 |
| chr1_-_12848720 | 0.26 |
ENST00000317869.7
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C like 1 |
| chr17_-_7263959 | 0.26 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr19_+_39125769 | 0.26 |
ENST00000602004.1
ENST00000599470.5 ENST00000321944.8 ENST00000593480.5 ENST00000358301.7 ENST00000593690.5 ENST00000599386.5 |
PAK4
|
p21 (RAC1) activated kinase 4 |
| chr5_+_150190035 | 0.26 |
ENST00000230671.7
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 member 7 |
| chr5_+_137867852 | 0.26 |
ENST00000421631.6
ENST00000239926.9 |
MYOT
|
myotilin |
| chr5_+_40679907 | 0.25 |
ENST00000302472.4
|
PTGER4
|
prostaglandin E receptor 4 |
| chr10_-_100185993 | 0.25 |
ENST00000421367.7
ENST00000370408.2 ENST00000407654.7 |
ERLIN1
|
ER lipid raft associated 1 |
| chr5_-_113294895 | 0.25 |
ENST00000514701.5
ENST00000302475.8 |
MCC
|
MCC regulator of WNT signaling pathway |
| chr4_+_70734419 | 0.25 |
ENST00000502653.5
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_+_8501807 | 0.25 |
ENST00000426878.2
ENST00000397386.7 ENST00000415597.5 ENST00000157600.8 |
LMCD1
|
LIM and cysteine rich domains 1 |
| chr14_+_94026314 | 0.25 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr20_-_34112205 | 0.25 |
ENST00000374980.3
|
EIF2S2
|
eukaryotic translation initiation factor 2 subunit beta |
| chr17_-_8162932 | 0.25 |
ENST00000488857.5
ENST00000316509.11 ENST00000481878.1 ENST00000498285.1 |
VAMP2
ENSG00000263620.1
|
vesicle associated membrane protein 2 novel protein |
| chr1_+_42825548 | 0.25 |
ENST00000372514.7
|
ERMAP
|
erythroblast membrane associated protein (Scianna blood group) |
| chr9_-_118417 | 0.25 |
ENST00000382500.4
|
FOXD4
|
forkhead box D4 |
| chr4_+_123396785 | 0.24 |
ENST00000505319.5
ENST00000651917.1 ENST00000610581.4 ENST00000339241.1 |
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr2_-_208129824 | 0.24 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C |
| chr5_+_145937793 | 0.24 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr7_+_92245960 | 0.24 |
ENST00000265742.8
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1 |
| chr17_-_35121487 | 0.24 |
ENST00000593039.5
|
ENSG00000267618.5
|
RAD51L3-RFFL readthrough |
| chr22_+_24607602 | 0.24 |
ENST00000447416.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_+_167248638 | 0.23 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr17_-_43546323 | 0.23 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr1_-_146021724 | 0.23 |
ENST00000475797.1
ENST00000497365.5 ENST00000336751.11 ENST00000634927.1 ENST00000421822.2 |
HJV
|
hemojuvelin BMP co-receptor |
| chr6_+_1609890 | 0.23 |
ENST00000645831.2
|
FOXC1
|
forkhead box C1 |
| chr6_+_27815010 | 0.23 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr12_-_27014300 | 0.23 |
ENST00000535819.1
ENST00000543803.5 ENST00000535423.5 ENST00000539741.5 ENST00000343028.9 ENST00000545600.1 ENST00000543088.5 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr11_+_67023085 | 0.23 |
ENST00000527043.6
|
SYT12
|
synaptotagmin 12 |
| chr11_+_307915 | 0.22 |
ENST00000616316.2
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr1_+_151766655 | 0.22 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr13_-_33285682 | 0.22 |
ENST00000336934.10
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr14_+_35122537 | 0.22 |
ENST00000604948.5
ENST00000321130.14 ENST00000534898.9 ENST00000605201.1 ENST00000250377.11 |
PRORP
|
protein only RNase P catalytic subunit |
| chr7_+_111091006 | 0.22 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr6_-_27814757 | 0.22 |
ENST00000333151.5
|
H2AC14
|
H2A clustered histone 14 |
| chr9_-_93453540 | 0.22 |
ENST00000375412.11
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr11_-_111913134 | 0.22 |
ENST00000533971.2
ENST00000526180.6 |
CRYAB
|
crystallin alpha B |
| chr2_-_25341886 | 0.22 |
ENST00000321117.10
|
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr16_-_4242068 | 0.21 |
ENST00000399609.7
|
SRL
|
sarcalumenin |
| chr1_+_149886906 | 0.21 |
ENST00000331380.4
|
H2AC20
|
H2A clustered histone 20 |
| chr7_+_111091119 | 0.21 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr19_+_42313374 | 0.21 |
ENST00000598766.1
ENST00000673187.1 |
TMEM145
|
transmembrane protein 145 |
| chr6_+_159169391 | 0.21 |
ENST00000297267.14
|
FNDC1
|
fibronectin type III domain containing 1 |
| chr19_+_4153616 | 0.21 |
ENST00000078445.7
ENST00000595923.5 ENST00000602257.5 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3 like 3 |
| chr11_-_111913195 | 0.21 |
ENST00000531198.5
ENST00000616970.5 ENST00000527899.6 |
CRYAB
|
crystallin alpha B |
| chr1_+_6034980 | 0.21 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr2_+_170178136 | 0.21 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.4 | 1.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 1.0 | GO:0060367 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.3 | 1.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.3 | 0.8 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.3 | 1.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 1.0 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.2 | 0.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 0.9 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.2 | 0.7 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 1.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 2.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 1.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.5 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.4 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.3 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 0.3 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 1.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 1.3 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.7 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.3 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.2 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 0.6 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.4 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 1.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 2.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 0.0 | 0.4 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0043397 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.5 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0072004 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0035498 | carnosine metabolic process(GO:0035498) |
| 0.0 | 0.6 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.2 | 1.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.4 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.0 | GO:0097546 | mediator complex(GO:0016592) ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 1.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 0.9 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 0.8 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.1 | 1.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.2 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.9 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |