Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
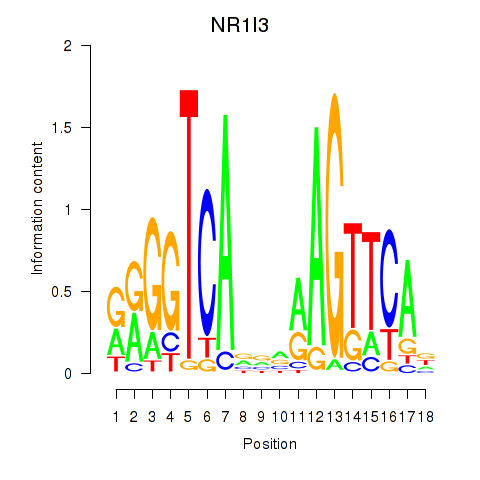
Results for NR1I3
Z-value: 0.33
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.12 | NR1I3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg38_v1_chr1_-_161238163_161238184 | -0.23 | 2.2e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_32439866 | 1.35 |
ENST00000374982.5
ENST00000395388.7 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chrX_-_15600953 | 0.58 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr9_-_72953047 | 0.47 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr12_+_53423849 | 0.45 |
ENST00000257863.9
ENST00000550311.5 ENST00000379791.7 |
AMHR2
|
anti-Mullerian hormone receptor type 2 |
| chr22_+_30607203 | 0.43 |
ENST00000407817.3
|
TCN2
|
transcobalamin 2 |
| chr1_-_19799872 | 0.40 |
ENST00000294543.11
|
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chr17_-_35943707 | 0.32 |
ENST00000615905.5
|
LYZL6
|
lysozyme like 6 |
| chr17_-_35943662 | 0.32 |
ENST00000618542.4
|
LYZL6
|
lysozyme like 6 |
| chr17_+_28506320 | 0.28 |
ENST00000579795.6
|
FOXN1
|
forkhead box N1 |
| chr6_-_56394094 | 0.28 |
ENST00000370819.5
|
COL21A1
|
collagen type XXI alpha 1 chain |
| chr7_-_73738831 | 0.25 |
ENST00000395147.9
ENST00000437775.7 |
ABHD11
|
abhydrolase domain containing 11 |
| chr11_+_63369779 | 0.24 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr7_-_73738792 | 0.23 |
ENST00000222800.8
ENST00000458339.6 |
ABHD11
|
abhydrolase domain containing 11 |
| chr12_-_108598036 | 0.23 |
ENST00000392806.4
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chrX_+_129738942 | 0.23 |
ENST00000371106.4
|
XPNPEP2
|
X-prolyl aminopeptidase 2 |
| chr6_+_33454543 | 0.19 |
ENST00000621915.1
ENST00000395064.3 |
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr16_+_30699155 | 0.18 |
ENST00000262518.9
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr1_-_23559490 | 0.18 |
ENST00000374561.6
|
ID3
|
inhibitor of DNA binding 3, HLH protein |
| chr16_-_16223467 | 0.17 |
ENST00000575728.1
ENST00000574094.6 ENST00000205557.12 |
ABCC6
|
ATP binding cassette subfamily C member 6 |
| chr1_+_32465046 | 0.16 |
ENST00000609129.2
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr17_-_40648646 | 0.16 |
ENST00000643318.1
|
SMARCE1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr1_+_14924100 | 0.15 |
ENST00000361144.9
|
KAZN
|
kazrin, periplakin interacting protein |
| chr16_+_53099100 | 0.14 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_151146643 | 0.13 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr3_-_115071333 | 0.12 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_151146611 | 0.12 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr17_-_64506616 | 0.11 |
ENST00000581697.5
ENST00000450599.7 ENST00000577922.6 |
DDX5
|
DEAD-box helicase 5 |
| chr17_-_29294141 | 0.11 |
ENST00000225388.9
|
NUFIP2
|
nuclear FMR1 interacting protein 2 |
| chr17_+_34255274 | 0.11 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr1_+_160127672 | 0.10 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr19_-_6604083 | 0.10 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr15_+_43593054 | 0.10 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr19_-_48637338 | 0.10 |
ENST00000601104.1
ENST00000222122.10 |
DBP
|
D-box binding PAR bZIP transcription factor |
| chr5_-_136193143 | 0.10 |
ENST00000607574.2
|
SMIM32
|
small integral membrane protein 32 |
| chr11_-_82900406 | 0.10 |
ENST00000313010.8
ENST00000393399.6 ENST00000680437.1 |
PRCP
|
prolylcarboxypeptidase |
| chr9_+_73151833 | 0.09 |
ENST00000456643.5
ENST00000257497.11 ENST00000415424.5 |
ANXA1
|
annexin A1 |
| chrX_+_70455093 | 0.09 |
ENST00000542398.1
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr20_+_6007245 | 0.09 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr19_-_7497447 | 0.09 |
ENST00000593942.5
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr17_-_8295342 | 0.09 |
ENST00000579192.5
ENST00000577745.2 |
SLC25A35
|
solute carrier family 25 member 35 |
| chr11_+_64241600 | 0.09 |
ENST00000535135.7
ENST00000652094.1 |
FKBP2
|
FKBP prolyl isomerase 2 |
| chr1_-_145918685 | 0.09 |
ENST00000369306.8
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr10_+_102503956 | 0.09 |
ENST00000369902.8
|
SUFU
|
SUFU negative regulator of hedgehog signaling |
| chr14_-_102509713 | 0.09 |
ENST00000286918.9
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr9_+_121286586 | 0.09 |
ENST00000545652.6
|
GSN
|
gelsolin |
| chr17_-_64506281 | 0.08 |
ENST00000225792.10
ENST00000585060.5 |
DDX5
|
DEAD-box helicase 5 |
| chr1_-_145918485 | 0.08 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr15_+_96332432 | 0.08 |
ENST00000559679.1
ENST00000394171.6 |
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr16_+_67873036 | 0.08 |
ENST00000358933.10
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr10_-_73808616 | 0.08 |
ENST00000299641.8
|
NDST2
|
N-deacetylase and N-sulfotransferase 2 |
| chr12_+_53380639 | 0.08 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr1_+_153357846 | 0.08 |
ENST00000368738.4
|
S100A9
|
S100 calcium binding protein A9 |
| chr18_-_21703688 | 0.07 |
ENST00000584464.1
ENST00000578270.5 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr15_+_96333111 | 0.07 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr4_+_70028452 | 0.07 |
ENST00000530128.5
ENST00000381057.3 ENST00000673563.1 |
HTN3
|
histatin 3 |
| chrX_-_111412162 | 0.07 |
ENST00000637570.1
ENST00000356220.8 ENST00000636035.2 ENST00000637453.1 ENST00000635795.1 |
DCX
|
doublecortin |
| chr3_-_116444983 | 0.07 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr8_+_11494367 | 0.06 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr2_-_39120383 | 0.06 |
ENST00000395038.6
|
SOS1
|
SOS Ras/Rac guanine nucleotide exchange factor 1 |
| chr2_-_86563470 | 0.06 |
ENST00000409225.2
|
CHMP3
|
charged multivesicular body protein 3 |
| chr17_+_64506953 | 0.06 |
ENST00000580188.1
ENST00000556440.7 ENST00000581056.5 |
CEP95
|
centrosomal protein 95 |
| chr14_+_22281097 | 0.06 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr9_+_131125973 | 0.06 |
ENST00000651639.1
ENST00000451030.5 ENST00000531584.1 |
NUP214
|
nucleoporin 214 |
| chr1_-_27356471 | 0.06 |
ENST00000486046.5
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr14_+_22508602 | 0.05 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr1_-_205321737 | 0.05 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2 |
| chr11_+_113908983 | 0.05 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr1_-_158426237 | 0.05 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr7_-_99735093 | 0.05 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr11_+_62789124 | 0.05 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr11_-_47848539 | 0.05 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160 |
| chr3_-_58433810 | 0.05 |
ENST00000474765.1
ENST00000485460.5 ENST00000302746.11 ENST00000383714.8 |
PDHB
|
pyruvate dehydrogenase E1 subunit beta |
| chr19_-_9858139 | 0.05 |
ENST00000590841.5
|
OLFM2
|
olfactomedin 2 |
| chr2_-_86563349 | 0.05 |
ENST00000409727.5
|
CHMP3
|
charged multivesicular body protein 3 |
| chr6_+_108166015 | 0.05 |
ENST00000368986.9
|
NR2E1
|
nuclear receptor subfamily 2 group E member 1 |
| chr3_+_122384167 | 0.04 |
ENST00000232125.9
ENST00000477892.5 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162 member A |
| chr19_-_45782388 | 0.04 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr7_-_150978284 | 0.04 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr5_-_177509814 | 0.04 |
ENST00000510898.7
ENST00000502885.5 ENST00000506493.5 |
DOK3
|
docking protein 3 |
| chr4_+_37244735 | 0.04 |
ENST00000309447.6
|
NWD2
|
NACHT and WD repeat domain containing 2 |
| chr16_+_527698 | 0.04 |
ENST00000219611.7
ENST00000562370.5 ENST00000568988.5 |
CAPN15
|
calpain 15 |
| chr17_+_46295099 | 0.04 |
ENST00000393465.7
ENST00000320254.5 |
LRRC37A
|
leucine rich repeat containing 37A |
| chr20_+_36461303 | 0.04 |
ENST00000475894.5
|
DLGAP4
|
DLG associated protein 4 |
| chr14_+_24130659 | 0.04 |
ENST00000267426.6
|
FITM1
|
fat storage inducing transmembrane protein 1 |
| chr11_-_57322197 | 0.03 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr2_-_86563382 | 0.03 |
ENST00000263856.9
|
CHMP3
|
charged multivesicular body protein 3 |
| chr3_-_56468346 | 0.03 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_+_27392612 | 0.03 |
ENST00000374024.4
|
GPR3
|
G protein-coupled receptor 3 |
| chr18_-_21704763 | 0.03 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr7_-_99784175 | 0.03 |
ENST00000651514.1
ENST00000336411.7 ENST00000415003.1 ENST00000354593.6 |
CYP3A4
|
cytochrome P450 family 3 subfamily A member 4 |
| chr11_-_47848467 | 0.03 |
ENST00000378460.6
|
NUP160
|
nucleoporin 160 |
| chr2_-_68952880 | 0.02 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr13_-_94479671 | 0.02 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr10_-_72088972 | 0.02 |
ENST00000317376.8
ENST00000412663.5 |
SPOCK2
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 2 |
| chr9_-_95317671 | 0.02 |
ENST00000490972.7
ENST00000647778.1 ENST00000649611.1 ENST00000289081.8 |
FANCC
|
FA complementation group C |
| chr11_-_47848313 | 0.02 |
ENST00000530326.5
ENST00000528071.5 |
NUP160
|
nucleoporin 160 |
| chr9_+_35605234 | 0.02 |
ENST00000336395.6
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr10_-_80172839 | 0.02 |
ENST00000265447.8
|
ANXA11
|
annexin A11 |
| chr20_-_46513516 | 0.02 |
ENST00000347606.8
ENST00000625284.2 ENST00000457685.6 |
ZNF334
|
zinc finger protein 334 |
| chrX_+_48786578 | 0.02 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1 |
| chr19_+_54200849 | 0.02 |
ENST00000626547.2
ENST00000302907.9 ENST00000391752.5 ENST00000402367.5 ENST00000391751.7 |
RPS9
|
ribosomal protein S9 |
| chr19_+_54201122 | 0.01 |
ENST00000391753.6
ENST00000441429.1 |
RPS9
|
ribosomal protein S9 |
| chr8_-_108787563 | 0.01 |
ENST00000297459.4
|
TMEM74
|
transmembrane protein 74 |
| chr9_+_35605277 | 0.01 |
ENST00000620767.4
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr1_+_43337803 | 0.01 |
ENST00000372470.9
ENST00000413998.7 |
MPL
|
MPL proto-oncogene, thrombopoietin receptor |
| chr1_-_155187272 | 0.01 |
ENST00000473363.3
|
ENSG00000273088.2
|
novel protein |
| chr5_-_177510348 | 0.01 |
ENST00000377112.8
ENST00000501403.6 ENST00000312943.10 |
DOK3
|
docking protein 3 |
| chr12_+_57604598 | 0.01 |
ENST00000548804.5
ENST00000550596.5 ENST00000551835.5 ENST00000549583.5 |
DTX3
|
deltex E3 ubiquitin ligase 3 |
| chr1_-_53220589 | 0.01 |
ENST00000294360.5
|
CZIB
|
CXXC motif containing zinc binding protein |
| chr14_+_52707192 | 0.01 |
ENST00000445930.7
ENST00000555339.5 ENST00000556813.1 |
PSMC6
|
proteasome 26S subunit, ATPase 6 |
| chr14_+_52707178 | 0.01 |
ENST00000612399.4
|
PSMC6
|
proteasome 26S subunit, ATPase 6 |
| chr6_-_119149124 | 0.01 |
ENST00000368475.8
|
FAM184A
|
family with sequence similarity 184 member A |
| chr11_-_64996963 | 0.01 |
ENST00000301887.9
ENST00000534177.1 |
BATF2
|
basic leucine zipper ATF-like transcription factor 2 |
| chr3_-_116445458 | 0.00 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr17_-_58219227 | 0.00 |
ENST00000581180.2
ENST00000313863.11 ENST00000393119.7 ENST00000678463.1 ENST00000676787.1 ENST00000580127.6 ENST00000585134.2 ENST00000581761.6 |
MKS1
|
MKS transition zone complex subunit 1 |
| chr6_-_34146080 | 0.00 |
ENST00000538487.7
ENST00000374181.8 |
GRM4
|
glutamate metabotropic receptor 4 |
| chr1_+_40817190 | 0.00 |
ENST00000443478.3
|
KCNQ4
|
potassium voltage-gated channel subfamily Q member 4 |
| chr16_-_66934144 | 0.00 |
ENST00000568572.5
|
CIAO2B
|
cytosolic iron-sulfur assembly component 2B |
| chr6_-_31541937 | 0.00 |
ENST00000456662.5
ENST00000431908.5 ENST00000456976.5 ENST00000428450.5 ENST00000418897.5 ENST00000396172.6 ENST00000419020.1 ENST00000428098.5 |
DDX39B
|
DExD-box helicase 39B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.6 | GO:0003051 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.3 | GO:1902232 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 0.5 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |