Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for NR2E1
Z-value: 0.76
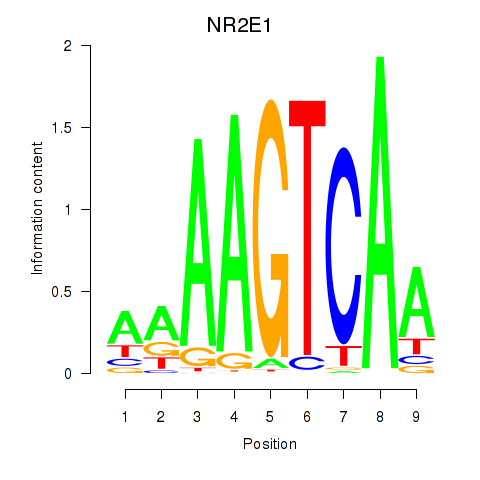
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.12 | NR2E1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg38_v1_chr6_+_108166015_108166034 | 0.26 | 1.7e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_31414694 | 2.70 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr1_-_205449924 | 2.42 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1 |
| chr5_-_73448769 | 2.25 |
ENST00000615637.3
|
FOXD1
|
forkhead box D1 |
| chr1_+_31576485 | 1.98 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr4_-_110198579 | 1.66 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_110198650 | 1.64 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr2_+_113127588 | 1.61 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr18_+_31447732 | 1.61 |
ENST00000257189.5
|
DSG3
|
desmoglein 3 |
| chr11_-_63671909 | 1.53 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr11_-_62556230 | 1.47 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_-_47981535 | 1.46 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr12_-_89352487 | 1.42 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr5_-_11588842 | 1.28 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr10_+_11742361 | 1.27 |
ENST00000379215.9
ENST00000420401.5 |
ECHDC3
|
enoyl-CoA hydratase domain containing 3 |
| chr5_-_11589019 | 1.24 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr7_+_116526277 | 1.19 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1 |
| chr3_+_122055355 | 1.16 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chrX_-_66040107 | 1.13 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr11_+_35176696 | 1.11 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_119803383 | 1.11 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chrX_-_66040072 | 1.10 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_66040057 | 1.08 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_66033664 | 1.07 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr17_-_43546323 | 1.04 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr22_-_30299482 | 1.00 |
ENST00000434291.5
|
ENSG00000248751.6
|
novel protein |
| chr6_+_12290353 | 0.99 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr11_+_35176575 | 0.98 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_+_56521951 | 0.98 |
ENST00000552247.6
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr7_-_107803215 | 0.97 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr12_+_56521990 | 0.96 |
ENST00000550726.5
ENST00000542360.1 |
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chrX_+_47585212 | 0.96 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr11_+_35176639 | 0.95 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_22508602 | 0.95 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr11_+_35176611 | 0.92 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chrX_+_136532205 | 0.91 |
ENST00000370634.8
|
VGLL1
|
vestigial like family member 1 |
| chr4_+_70197924 | 0.89 |
ENST00000514097.5
|
ODAM
|
odontogenic, ameloblast associated |
| chr12_-_27971970 | 0.86 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr1_-_162412117 | 0.85 |
ENST00000367929.3
|
SH2D1B
|
SH2 domain containing 1B |
| chr8_-_17002327 | 0.83 |
ENST00000180166.6
|
FGF20
|
fibroblast growth factor 20 |
| chr9_-_96302104 | 0.82 |
ENST00000375262.4
ENST00000650386.1 |
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr2_-_89143133 | 0.79 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr17_+_2056073 | 0.78 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr12_-_94650506 | 0.77 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr2_-_207165923 | 0.76 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr17_+_17179527 | 0.76 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr6_+_106098933 | 0.74 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr12_-_81758665 | 0.74 |
ENST00000549325.5
ENST00000550584.6 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr4_-_4542034 | 0.73 |
ENST00000306200.7
|
STX18
|
syntaxin 18 |
| chr11_-_111923722 | 0.73 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr9_-_21994345 | 0.69 |
ENST00000579755.2
ENST00000530628.2 |
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr1_-_197067234 | 0.68 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr14_+_66824439 | 0.68 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr3_+_136957948 | 0.67 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr2_+_28395511 | 0.64 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr9_-_96302142 | 0.63 |
ENST00000648799.1
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr12_-_91178520 | 0.62 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr12_+_53268289 | 0.62 |
ENST00000257934.9
|
ESPL1
|
extra spindle pole bodies like 1, separase |
| chr2_-_213150236 | 0.61 |
ENST00000442445.1
ENST00000342002.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr2_+_114442616 | 0.61 |
ENST00000410059.6
|
DPP10
|
dipeptidyl peptidase like 10 |
| chrX_-_49184789 | 0.61 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr11_-_128587551 | 0.60 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr11_-_31812171 | 0.60 |
ENST00000639409.1
ENST00000640975.1 |
PAX6
|
paired box 6 |
| chr3_+_128052390 | 0.59 |
ENST00000481210.5
ENST00000243253.8 |
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr2_+_191276885 | 0.59 |
ENST00000392316.5
|
MYO1B
|
myosin IB |
| chr1_+_165895564 | 0.59 |
ENST00000469256.6
|
UCK2
|
uridine-cytidine kinase 2 |
| chr2_+_11539833 | 0.58 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr5_-_140633690 | 0.57 |
ENST00000512545.1
ENST00000401743.6 |
CD14
|
CD14 molecule |
| chr3_+_141387616 | 0.56 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_39955112 | 0.56 |
ENST00000420632.6
ENST00000372811.10 ENST00000434861.5 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr12_-_52493250 | 0.56 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr8_+_11809135 | 0.55 |
ENST00000528643.5
ENST00000525777.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_-_243850216 | 0.55 |
ENST00000673466.1
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr9_-_96302170 | 0.55 |
ENST00000375263.8
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr1_-_154183199 | 0.55 |
ENST00000323144.12
ENST00000330188.13 ENST00000328159.9 ENST00000611659.4 |
TPM3
|
tropomyosin 3 |
| chr2_+_90038848 | 0.54 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr5_-_179620933 | 0.53 |
ENST00000521173.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_-_116444983 | 0.53 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr3_+_189631373 | 0.52 |
ENST00000264731.8
ENST00000418709.6 ENST00000320472.9 ENST00000392460.7 ENST00000440651.6 |
TP63
|
tumor protein p63 |
| chr1_-_99766620 | 0.51 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr6_-_87702221 | 0.51 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chr3_-_33645433 | 0.50 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_+_116170742 | 0.50 |
ENST00000371906.5
ENST00000681852.1 |
AGTR2
|
angiotensin II receptor type 2 |
| chr10_-_68407242 | 0.50 |
ENST00000399200.7
ENST00000602465.6 ENST00000454950.6 |
RUFY2
|
RUN and FYVE domain containing 2 |
| chr2_-_55296361 | 0.50 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr20_+_1895365 | 0.49 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha |
| chr7_+_116672357 | 0.49 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr4_-_142846275 | 0.49 |
ENST00000513000.5
ENST00000509777.5 ENST00000503927.5 |
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_+_213989691 | 0.48 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr19_+_11435619 | 0.47 |
ENST00000589126.5
ENST00000588269.1 ENST00000587509.5 ENST00000591462.6 ENST00000592741.5 ENST00000677123.1 ENST00000593101.5 ENST00000587327.5 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr16_+_3018390 | 0.46 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr2_+_230759918 | 0.45 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr3_-_197183806 | 0.45 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr4_-_156970903 | 0.44 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr3_+_12351493 | 0.44 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr17_+_67377272 | 0.43 |
ENST00000581322.6
ENST00000299954.13 |
PITPNC1
|
phosphatidylinositol transfer protein cytoplasmic 1 |
| chr3_-_185821092 | 0.43 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr19_-_14848922 | 0.43 |
ENST00000641129.1
|
OR7A10
|
olfactory receptor family 7 subfamily A member 10 |
| chrX_-_54798253 | 0.43 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr3_+_12351470 | 0.42 |
ENST00000287820.10
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chrX_+_47218670 | 0.42 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr10_-_68407189 | 0.42 |
ENST00000388768.6
|
RUFY2
|
RUN and FYVE domain containing 2 |
| chr2_-_157444044 | 0.42 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr6_+_42564060 | 0.42 |
ENST00000372903.6
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr4_+_95091462 | 0.41 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr11_-_31807617 | 0.40 |
ENST00000639920.1
ENST00000640460.1 |
PAX6
|
paired box 6 |
| chr1_-_154183130 | 0.40 |
ENST00000368531.6
ENST00000368533.8 |
TPM3
|
tropomyosin 3 |
| chr6_-_32151899 | 0.40 |
ENST00000211413.10
|
PRRT1
|
proline rich transmembrane protein 1 |
| chr15_-_56245074 | 0.39 |
ENST00000674082.1
|
RFX7
|
regulatory factor X7 |
| chr10_+_122560751 | 0.38 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr17_+_37375974 | 0.38 |
ENST00000615133.2
ENST00000611038.4 |
C17orf78
|
chromosome 17 open reading frame 78 |
| chr3_+_101827982 | 0.38 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr1_+_76867469 | 0.38 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr5_-_88883199 | 0.38 |
ENST00000514015.5
ENST00000503075.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr9_-_92482350 | 0.38 |
ENST00000375543.2
|
ASPN
|
asporin |
| chrX_-_31178220 | 0.38 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr6_+_125219804 | 0.38 |
ENST00000524679.1
|
TPD52L1
|
TPD52 like 1 |
| chr17_-_42681840 | 0.37 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr5_-_83673544 | 0.37 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_115029823 | 0.37 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr7_-_13986439 | 0.37 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr1_+_50109817 | 0.36 |
ENST00000652353.1
ENST00000371821.6 ENST00000652274.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chrX_-_53684119 | 0.36 |
ENST00000342160.7
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1 |
| chr10_+_122560679 | 0.36 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_7936177 | 0.36 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chrX_-_31178149 | 0.35 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr14_-_56810448 | 0.35 |
ENST00000339475.10
ENST00000555006.5 ENST00000672264.2 ENST00000554559.5 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr8_-_92017292 | 0.35 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr16_+_21678514 | 0.35 |
ENST00000286149.8
ENST00000388958.8 |
OTOA
|
otoancorin |
| chr12_-_10172117 | 0.35 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr5_-_113434978 | 0.34 |
ENST00000390666.4
|
TSSK1B
|
testis specific serine kinase 1B |
| chr15_+_67067780 | 0.34 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chrX_+_47218232 | 0.34 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr12_-_99154867 | 0.34 |
ENST00000549025.6
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_24460630 | 0.34 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr10_+_122560639 | 0.34 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr8_+_11704151 | 0.33 |
ENST00000526716.5
ENST00000532059.6 ENST00000335135.8 ENST00000622443.3 |
GATA4
|
GATA binding protein 4 |
| chr12_+_14365729 | 0.33 |
ENST00000536444.5
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr19_-_633500 | 0.33 |
ENST00000588649.7
|
POLRMT
|
RNA polymerase mitochondrial |
| chr18_+_24460655 | 0.33 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chrX_-_33128360 | 0.33 |
ENST00000378677.6
|
DMD
|
dystrophin |
| chr1_-_154178174 | 0.33 |
ENST00000302206.9
|
TPM3
|
tropomyosin 3 |
| chr6_-_130222813 | 0.33 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr2_-_208146150 | 0.33 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr9_+_96928843 | 0.33 |
ENST00000372322.4
|
NUTM2G
|
NUT family member 2G |
| chr19_-_46717076 | 0.33 |
ENST00000601806.5
ENST00000593363.1 ENST00000291281.9 ENST00000598633.1 ENST00000595515.5 ENST00000433867.5 |
PRKD2
|
protein kinase D2 |
| chr5_-_151157722 | 0.32 |
ENST00000517486.5
ENST00000377751.9 ENST00000521512.5 ENST00000517757.5 ENST00000354546.10 |
ANXA6
|
annexin A6 |
| chr6_-_155455830 | 0.32 |
ENST00000159060.3
|
NOX3
|
NADPH oxidase 3 |
| chr16_-_20697680 | 0.32 |
ENST00000520010.6
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr4_+_80266639 | 0.31 |
ENST00000456523.3
ENST00000312465.12 |
FGF5
|
fibroblast growth factor 5 |
| chr3_-_112133218 | 0.31 |
ENST00000488580.5
ENST00000308910.9 ENST00000460387.6 ENST00000484193.5 ENST00000487901.1 |
GCSAM
|
germinal center associated signaling and motility |
| chr16_-_69754913 | 0.30 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr8_-_123396412 | 0.30 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr4_-_152382522 | 0.30 |
ENST00000296555.11
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr12_-_91153149 | 0.30 |
ENST00000550758.1
|
DCN
|
decorin |
| chr12_+_14365661 | 0.30 |
ENST00000261168.9
ENST00000538511.5 ENST00000545723.1 ENST00000543189.5 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_-_243843164 | 0.30 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr14_+_51489112 | 0.30 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6 |
| chr19_-_46654657 | 0.29 |
ENST00000300875.4
|
DACT3
|
dishevelled binding antagonist of beta catenin 3 |
| chr3_-_114178698 | 0.29 |
ENST00000467632.5
|
DRD3
|
dopamine receptor D3 |
| chr17_-_676348 | 0.29 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr6_+_54307856 | 0.29 |
ENST00000370869.7
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr3_-_58211212 | 0.28 |
ENST00000461914.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3 |
| chr3_-_132037800 | 0.28 |
ENST00000617767.4
|
CPNE4
|
copine 4 |
| chr9_-_94328644 | 0.28 |
ENST00000341207.5
ENST00000253262.9 |
NUTM2F
|
NUT family member 2F |
| chr1_-_20508095 | 0.28 |
ENST00000264198.5
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr7_+_16526824 | 0.27 |
ENST00000401542.3
|
LRRC72
|
leucine rich repeat containing 72 |
| chr5_-_16936231 | 0.27 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr20_+_43945677 | 0.27 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr1_+_62597510 | 0.27 |
ENST00000371129.4
|
ANGPTL3
|
angiopoietin like 3 |
| chr1_-_115768702 | 0.27 |
ENST00000261448.6
|
CASQ2
|
calsequestrin 2 |
| chr1_+_67685342 | 0.27 |
ENST00000617962.2
|
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr10_-_68232108 | 0.27 |
ENST00000373673.5
|
ATOH7
|
atonal bHLH transcription factor 7 |
| chr5_+_32711723 | 0.27 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor 3 |
| chr4_+_88592426 | 0.27 |
ENST00000431413.5
ENST00000402738.6 ENST00000422770.5 ENST00000407637.5 |
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr9_-_92482499 | 0.27 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr7_-_106285898 | 0.26 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr7_+_107168961 | 0.26 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr18_+_44680093 | 0.26 |
ENST00000426838.8
ENST00000677068.1 |
SETBP1
|
SET binding protein 1 |
| chr11_-_111910790 | 0.26 |
ENST00000533280.6
|
CRYAB
|
crystallin alpha B |
| chr7_-_108240049 | 0.25 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr10_+_84230660 | 0.25 |
ENST00000652073.1
|
RGR
|
retinal G protein coupled receptor |
| chr16_+_7304219 | 0.25 |
ENST00000675562.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr1_+_50105666 | 0.25 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr20_+_64164474 | 0.25 |
ENST00000622439.4
ENST00000536311.5 |
MYT1
|
myelin transcription factor 1 |
| chr1_-_155978144 | 0.24 |
ENST00000313695.11
ENST00000497907.5 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr20_+_64164446 | 0.24 |
ENST00000328439.6
|
MYT1
|
myelin transcription factor 1 |
| chr16_-_58546702 | 0.24 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr11_-_111910830 | 0.24 |
ENST00000526167.5
ENST00000651650.1 |
CRYAB
|
crystallin alpha B |
| chr2_-_55010348 | 0.24 |
ENST00000394609.6
|
RTN4
|
reticulon 4 |
| chr7_-_42152396 | 0.24 |
ENST00000642432.1
ENST00000647255.1 ENST00000677288.1 |
GLI3
|
GLI family zinc finger 3 |
| chr10_-_24721866 | 0.24 |
ENST00000416305.1
ENST00000320481.10 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_-_42152444 | 0.23 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3 |
| chr20_+_64164566 | 0.23 |
ENST00000650655.1
|
MYT1
|
myelin transcription factor 1 |
| chr14_+_61697622 | 0.23 |
ENST00000539097.2
|
HIF1A
|
hypoxia inducible factor 1 subunit alpha |
| chr10_-_21174187 | 0.23 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr16_-_3018170 | 0.22 |
ENST00000572154.1
ENST00000328796.5 |
CLDN6
|
claudin 6 |
| chr9_-_16727980 | 0.22 |
ENST00000418777.5
ENST00000468187.6 |
BNC2
|
basonuclin 2 |
| chr3_-_50322759 | 0.22 |
ENST00000442581.1
ENST00000447092.5 |
HYAL2
|
hyaluronidase 2 |
| chr11_-_13496018 | 0.22 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr19_-_45730861 | 0.22 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 2.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.4 | 1.2 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.3 | 1.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.3 | 3.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.3 | 3.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 1.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.7 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.7 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.7 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 4.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 2.0 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 0.6 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.2 | 0.7 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.2 | 0.5 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 0.5 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.5 | GO:0060873 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.1 | 0.6 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.9 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.6 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.6 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.5 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.6 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.8 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.3 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 1.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.2 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.1 | GO:0060129 | mesodermal cell migration(GO:0008078) corticotropin hormone secreting cell differentiation(GO:0060128) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.3 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.6 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.6 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.4 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 1.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.0 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of phospholipase activity(GO:0010519) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.3 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.8 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.6 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.3 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 1.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.4 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.4 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.0 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 0.8 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 1.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 3.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.5 | 1.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.4 | 1.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.3 | 3.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.3 | 2.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.5 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 4.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.6 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 2.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.5 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 2.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 1.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |