Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
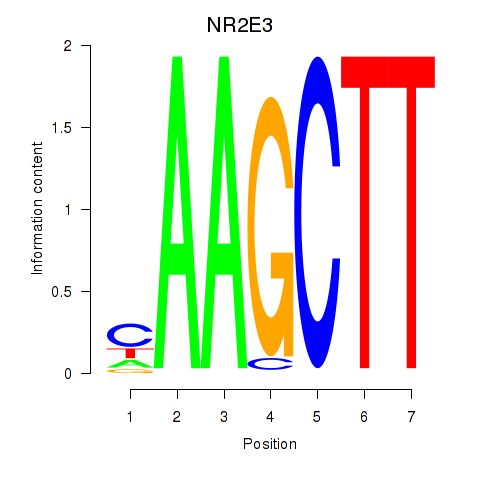
Results for NR2E3
Z-value: 0.55
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000278570.5 | NR2E3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E3 | hg38_v1_chr15_+_71810539_71810592 | 0.25 | 1.9e-01 | Click! |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_215393126 | 2.84 |
ENST00000456923.5
|
FN1
|
fibronectin 1 |
| chr3_-_195583931 | 1.14 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr9_+_33795551 | 0.96 |
ENST00000379405.4
|
PRSS3
|
serine protease 3 |
| chrX_-_133753681 | 0.75 |
ENST00000406757.2
|
GPC3
|
glypican 3 |
| chr8_+_53851786 | 0.71 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr7_+_142770960 | 0.67 |
ENST00000632805.1
ENST00000633969.1 ENST00000539842.6 |
PRSS2
|
serine protease 2 |
| chr2_-_55296361 | 0.66 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr8_+_40153475 | 0.65 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator |
| chr20_-_57711536 | 0.61 |
ENST00000265626.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_+_142760398 | 0.59 |
ENST00000632998.1
|
PRSS2
|
serine protease 2 |
| chr6_+_73695779 | 0.56 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr4_-_158173004 | 0.50 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr1_+_152514474 | 0.49 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1 |
| chr11_+_43942627 | 0.45 |
ENST00000617612.3
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr6_+_130018565 | 0.44 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr2_+_101839815 | 0.44 |
ENST00000421882.5
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_-_207166818 | 0.44 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr3_-_33645253 | 0.43 |
ENST00000333778.10
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr18_+_58045683 | 0.42 |
ENST00000592846.5
ENST00000675801.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr14_+_78403686 | 0.42 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr6_-_4135459 | 0.41 |
ENST00000495548.1
ENST00000380125.6 ENST00000465828.5 ENST00000380118.8 ENST00000361538.6 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr17_-_66192125 | 0.41 |
ENST00000535342.7
|
CEP112
|
centrosomal protein 112 |
| chr11_+_77034392 | 0.39 |
ENST00000528622.5
ENST00000622824.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 |
| chr2_-_207167220 | 0.38 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr10_-_14330879 | 0.37 |
ENST00000357447.7
|
FRMD4A
|
FERM domain containing 4A |
| chr10_-_101843765 | 0.37 |
ENST00000370046.5
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr4_-_109801978 | 0.36 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr2_+_190927649 | 0.36 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr12_+_93569814 | 0.35 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr12_-_119804298 | 0.35 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr12_-_119804472 | 0.34 |
ENST00000678087.1
ENST00000677993.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr15_-_55917080 | 0.34 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr15_+_73684731 | 0.33 |
ENST00000560995.5
|
CD276
|
CD276 molecule |
| chr3_+_189789734 | 0.32 |
ENST00000437221.5
ENST00000392463.6 ENST00000392461.7 ENST00000449992.5 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr11_+_46381753 | 0.32 |
ENST00000407067.1
|
MDK
|
midkine |
| chr18_+_58149314 | 0.32 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_+_32502114 | 0.31 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr18_+_58045642 | 0.31 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_+_46381698 | 0.31 |
ENST00000395565.5
|
MDK
|
midkine |
| chr6_+_37929959 | 0.31 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr12_-_66678934 | 0.31 |
ENST00000545666.5
ENST00000398016.7 ENST00000359742.9 ENST00000538211.5 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_-_111488795 | 0.29 |
ENST00000472933.2
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr7_+_18495723 | 0.29 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr1_-_149936324 | 0.28 |
ENST00000369140.7
|
MTMR11
|
myotubularin related protein 11 |
| chr1_-_153375591 | 0.28 |
ENST00000368737.5
|
S100A12
|
S100 calcium binding protein A12 |
| chr12_+_10505890 | 0.28 |
ENST00000538173.1
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B |
| chr16_-_46621345 | 0.27 |
ENST00000303383.8
|
SHCBP1
|
SHC binding and spindle associated 1 |
| chr4_-_167234552 | 0.27 |
ENST00000512648.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr3_+_69866217 | 0.27 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr7_+_142749465 | 0.27 |
ENST00000486171.5
ENST00000619214.4 ENST00000311737.12 |
PRSS1
|
serine protease 1 |
| chr5_-_22853320 | 0.26 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chrX_-_107716401 | 0.26 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr12_-_89352487 | 0.26 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr9_+_136952896 | 0.26 |
ENST00000371632.7
|
LCN12
|
lipocalin 12 |
| chr12_+_20695553 | 0.26 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr3_-_71360753 | 0.25 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr3_+_57890011 | 0.25 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr5_+_134525649 | 0.25 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr3_+_160677152 | 0.25 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr2_-_65366650 | 0.25 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr1_-_94925759 | 0.25 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr2_+_33134579 | 0.25 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_165827574 | 0.25 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_-_111563934 | 0.24 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr1_+_15764419 | 0.24 |
ENST00000441801.6
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_+_33134620 | 0.24 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr20_+_33237712 | 0.24 |
ENST00000618484.1
|
BPIFA1
|
BPI fold containing family A member 1 |
| chr3_-_71306012 | 0.24 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr8_-_61646807 | 0.24 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr9_+_100578071 | 0.24 |
ENST00000307584.6
|
CAVIN4
|
caveolae associated protein 4 |
| chr6_-_32184227 | 0.23 |
ENST00000450110.5
ENST00000375067.7 ENST00000375056.6 |
AGER
|
advanced glycosylation end-product specific receptor |
| chr15_+_67138001 | 0.23 |
ENST00000439724.7
|
SMAD3
|
SMAD family member 3 |
| chr16_+_50266530 | 0.23 |
ENST00000566433.6
ENST00000394697.7 ENST00000673801.1 |
ADCY7
|
adenylate cyclase 7 |
| chrX_+_17737443 | 0.23 |
ENST00000398080.5
ENST00000380045.7 ENST00000380043.7 ENST00000380041.8 |
SCML1
|
Scm polycomb group protein like 1 |
| chr4_+_188139438 | 0.22 |
ENST00000332517.4
|
TRIML1
|
tripartite motif family like 1 |
| chr19_-_42594918 | 0.22 |
ENST00000244336.10
|
CEACAM8
|
CEA cell adhesion molecule 8 |
| chr15_-_55917129 | 0.21 |
ENST00000338963.6
ENST00000508342.5 |
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr4_-_20984011 | 0.21 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr20_-_14337602 | 0.21 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr22_-_28712136 | 0.21 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr4_+_95840084 | 0.21 |
ENST00000295266.6
|
PDHA2
|
pyruvate dehydrogenase E1 subunit alpha 2 |
| chr6_-_130970428 | 0.21 |
ENST00000529208.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr15_-_56245074 | 0.21 |
ENST00000674082.1
|
RFX7
|
regulatory factor X7 |
| chr17_-_40994159 | 0.21 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr6_+_21593742 | 0.21 |
ENST00000244745.4
|
SOX4
|
SRY-box transcription factor 4 |
| chr1_+_22653228 | 0.21 |
ENST00000509305.6
|
C1QB
|
complement C1q B chain |
| chr5_-_151093566 | 0.19 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr3_+_188153271 | 0.19 |
ENST00000448637.5
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_63839005 | 0.19 |
ENST00000508192.5
ENST00000361128.9 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr6_+_146029059 | 0.19 |
ENST00000282753.6
|
GRM1
|
glutamate metabotropic receptor 1 |
| chr3_+_141386393 | 0.19 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_-_58577367 | 0.19 |
ENST00000464064.5
ENST00000360997.7 |
FAM107A
|
family with sequence similarity 107 member A |
| chr16_+_30762289 | 0.19 |
ENST00000566811.5
ENST00000565995.5 ENST00000563683.5 ENST00000357890.9 ENST00000324685.11 ENST00000565931.1 |
RNF40
|
ring finger protein 40 |
| chr11_+_63839173 | 0.18 |
ENST00000502399.7
ENST00000425897.3 ENST00000513765.7 ENST00000679216.1 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr17_-_8152380 | 0.18 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1 |
| chr11_+_63839086 | 0.18 |
ENST00000350490.11
ENST00000402010.8 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr5_-_88731827 | 0.18 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chrX_+_79144664 | 0.18 |
ENST00000645147.2
|
GPR174
|
G protein-coupled receptor 174 |
| chr9_+_70043840 | 0.18 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr9_-_39288138 | 0.18 |
ENST00000297668.10
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr19_-_14518383 | 0.17 |
ENST00000254322.3
ENST00000595139.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr3_-_33645433 | 0.17 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_+_36166556 | 0.17 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2 |
| chr22_+_23763207 | 0.17 |
ENST00000305199.9
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr10_-_114144599 | 0.17 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr13_-_33205997 | 0.17 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr7_-_101217569 | 0.17 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chrX_-_139832235 | 0.17 |
ENST00000327569.7
ENST00000361648.6 |
ATP11C
|
ATPase phospholipid transporting 11C |
| chr18_+_3448456 | 0.17 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr4_-_76007501 | 0.17 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr5_-_11589019 | 0.17 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr17_-_64130813 | 0.16 |
ENST00000606895.2
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr6_-_33711717 | 0.16 |
ENST00000374214.3
|
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr3_-_58577648 | 0.16 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A |
| chr6_+_26183750 | 0.16 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr17_-_66191855 | 0.16 |
ENST00000583358.1
ENST00000392769.6 |
CEP112
|
centrosomal protein 112 |
| chr7_+_107168961 | 0.16 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr6_-_90587018 | 0.16 |
ENST00000369332.7
ENST00000369329.8 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr5_-_16738341 | 0.16 |
ENST00000515803.5
|
MYO10
|
myosin X |
| chr17_-_59155235 | 0.16 |
ENST00000581068.5
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_-_197297523 | 0.16 |
ENST00000434148.1
ENST00000412364.2 ENST00000661013.1 ENST00000666007.1 ENST00000422288.6 ENST00000456699.6 ENST00000392380.6 ENST00000670935.1 ENST00000656087.1 ENST00000436682.6 ENST00000662727.1 ENST00000670455.1 ENST00000659221.1 ENST00000671185.1 ENST00000669565.1 ENST00000660898.1 ENST00000667971.1 ENST00000661453.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr1_-_111563956 | 0.16 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr20_+_43945677 | 0.16 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr7_-_84194781 | 0.15 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A |
| chr12_-_99154746 | 0.15 |
ENST00000549558.6
ENST00000550693.6 ENST00000549493.6 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr22_-_41285868 | 0.15 |
ENST00000422838.1
ENST00000405486.5 |
RANGAP1
|
Ran GTPase activating protein 1 |
| chr19_+_3762705 | 0.15 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr18_+_7754959 | 0.15 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M |
| chr3_-_71582096 | 0.15 |
ENST00000648895.1
|
FOXP1
|
forkhead box P1 |
| chr21_-_26171110 | 0.15 |
ENST00000359726.7
|
APP
|
amyloid beta precursor protein |
| chr1_+_66534107 | 0.15 |
ENST00000371037.9
ENST00000684651.1 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chr1_-_120054225 | 0.15 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr5_+_169637241 | 0.15 |
ENST00000520908.7
|
DOCK2
|
dedicator of cytokinesis 2 |
| chr13_-_51453015 | 0.15 |
ENST00000442263.4
ENST00000311234.9 |
INTS6
|
integrator complex subunit 6 |
| chr16_+_68644988 | 0.15 |
ENST00000429102.6
|
CDH3
|
cadherin 3 |
| chr5_-_43557689 | 0.15 |
ENST00000338972.8
ENST00000511321.5 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr1_+_66534171 | 0.14 |
ENST00000682762.1
ENST00000424320.6 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chr5_+_141355003 | 0.14 |
ENST00000571252.3
ENST00000612927.1 |
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr17_+_59155726 | 0.14 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr2_-_222298659 | 0.14 |
ENST00000336840.11
ENST00000344493.9 |
PAX3
|
paired box 3 |
| chr9_-_137590485 | 0.14 |
ENST00000298585.3
|
ZMYND19
|
zinc finger MYND-type containing 19 |
| chr8_+_11808417 | 0.14 |
ENST00000525954.5
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr9_-_21482313 | 0.14 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr14_+_73569115 | 0.14 |
ENST00000622407.4
ENST00000238651.10 |
ACOT2
|
acyl-CoA thioesterase 2 |
| chr10_+_74176537 | 0.14 |
ENST00000672394.1
|
ADK
|
adenosine kinase |
| chr10_+_69278492 | 0.14 |
ENST00000643399.2
|
HK1
|
hexokinase 1 |
| chr5_-_170389634 | 0.14 |
ENST00000521859.1
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr5_-_95682968 | 0.14 |
ENST00000274432.13
|
SPATA9
|
spermatogenesis associated 9 |
| chr12_+_78863962 | 0.14 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1 |
| chr19_+_3762665 | 0.14 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr16_+_68645290 | 0.14 |
ENST00000264012.9
|
CDH3
|
cadherin 3 |
| chr15_-_58014097 | 0.14 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr12_+_20695323 | 0.14 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr1_+_84164684 | 0.14 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_162242998 | 0.13 |
ENST00000627638.2
ENST00000447386.5 |
FAP
|
fibroblast activation protein alpha |
| chr10_-_89643870 | 0.13 |
ENST00000322191.10
ENST00000342512.3 |
PANK1
|
pantothenate kinase 1 |
| chr12_-_10998304 | 0.13 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr1_+_84164962 | 0.13 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr16_-_15055969 | 0.13 |
ENST00000566419.1
ENST00000568320.5 |
NTAN1
|
N-terminal asparagine amidase |
| chr3_+_141324208 | 0.13 |
ENST00000509842.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr21_-_30216047 | 0.13 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr19_-_14517425 | 0.13 |
ENST00000676577.1
ENST00000677204.1 ENST00000598235.2 |
DNAJB1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr5_+_96743536 | 0.13 |
ENST00000515663.5
|
CAST
|
calpastatin |
| chr10_-_70170466 | 0.13 |
ENST00000373239.2
ENST00000373241.9 ENST00000373242.6 |
SAR1A
|
secretion associated Ras related GTPase 1A |
| chrX_+_12906639 | 0.13 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr18_+_44680875 | 0.13 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr14_+_73569266 | 0.13 |
ENST00000613168.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr4_+_159241016 | 0.13 |
ENST00000644902.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr4_-_145938775 | 0.13 |
ENST00000508784.6
|
ZNF827
|
zinc finger protein 827 |
| chr18_-_74457944 | 0.13 |
ENST00000400291.2
|
DIPK1C
|
divergent protein kinase domain 1C |
| chr1_+_203626775 | 0.12 |
ENST00000367218.7
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr1_+_84164370 | 0.12 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_-_101846957 | 0.12 |
ENST00000338858.9
|
OLFM3
|
olfactomedin 3 |
| chr2_-_287687 | 0.12 |
ENST00000401489.6
ENST00000619265.4 |
ALKAL2
|
ALK and LTK ligand 2 |
| chr9_+_126613922 | 0.12 |
ENST00000526117.6
ENST00000373474.9 ENST00000355497.10 |
LMX1B
|
LIM homeobox transcription factor 1 beta |
| chr6_+_69232406 | 0.12 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr3_-_71305986 | 0.12 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr7_-_29195186 | 0.12 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr11_+_30231000 | 0.12 |
ENST00000254122.8
ENST00000533718.2 ENST00000417547.1 |
FSHB
|
follicle stimulating hormone subunit beta |
| chr6_-_127918540 | 0.12 |
ENST00000368250.5
|
THEMIS
|
thymocyte selection associated |
| chr2_-_210303608 | 0.12 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr4_+_3369828 | 0.12 |
ENST00000338806.4
|
RGS12
|
regulator of G protein signaling 12 |
| chr15_+_65622009 | 0.12 |
ENST00000544319.6
|
SLC24A1
|
solute carrier family 24 member 1 |
| chr7_-_151083400 | 0.12 |
ENST00000482202.5
ENST00000297533.9 |
TMUB1
|
transmembrane and ubiquitin like domain containing 1 |
| chr17_+_77287891 | 0.12 |
ENST00000449803.6
ENST00000431235.6 |
SEPTIN9
|
septin 9 |
| chr11_-_83034195 | 0.12 |
ENST00000531021.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr15_-_55270280 | 0.12 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr8_+_36784324 | 0.12 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr3_-_186544377 | 0.11 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr12_-_70920612 | 0.11 |
ENST00000283228.7
|
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr3_-_114179052 | 0.11 |
ENST00000383673.5
ENST00000295881.9 |
DRD3
|
dopamine receptor D3 |
| chr8_-_94436926 | 0.11 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_-_205935822 | 0.11 |
ENST00000340781.8
|
SLC26A9
|
solute carrier family 26 member 9 |
| chrX_+_68829009 | 0.11 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr15_-_58279245 | 0.11 |
ENST00000558231.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr11_-_82846128 | 0.11 |
ENST00000679809.1
ENST00000680186.1 ENST00000681592.1 |
PRCP
|
prolylcarboxypeptidase |
| chr1_-_217631034 | 0.11 |
ENST00000366934.3
ENST00000366935.8 |
GPATCH2
|
G-patch domain containing 2 |
| chr19_+_41363989 | 0.11 |
ENST00000413014.6
|
TMEM91
|
transmembrane protein 91 |
| chr1_+_148889403 | 0.11 |
ENST00000464103.5
ENST00000534536.5 ENST00000369356.8 ENST00000369354.7 ENST00000369347.8 ENST00000369349.7 ENST00000369351.7 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_66534082 | 0.11 |
ENST00000683257.1
ENST00000684083.1 ENST00000682938.1 ENST00000683581.1 ENST00000682293.1 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.4 | 1.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 0.6 | GO:0019089 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.1 | 0.8 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.6 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:1905204 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:2000670 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0042946 | glucoside transport(GO:0042946) |
| 0.0 | 1.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.0 | 0.1 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) sensory system development(GO:0048880) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0035483 | negative regulation of sodium:proton antiporter activity(GO:0032416) gastric motility(GO:0035482) gastric emptying(GO:0035483) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.5 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.4 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.2 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 3.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0050211 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |