Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for OLIG1
Z-value: 0.61
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.13 | OLIG1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OLIG1 | hg38_v1_chr21_+_33070133_33070149 | 0.08 | 6.8e-01 | Click! |
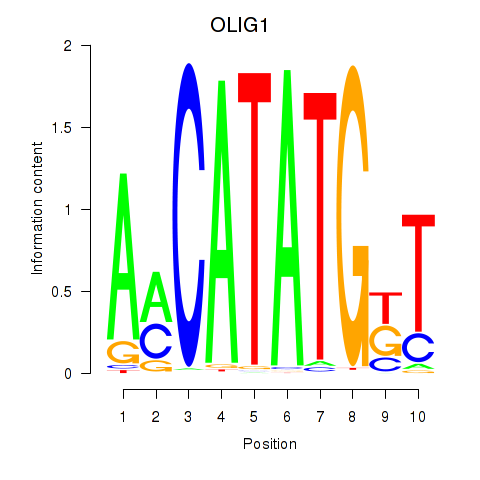
Activity profile of OLIG1 motif
Sorted Z-values of OLIG1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_68447453 | 3.77 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr1_-_153094521 | 3.01 |
ENST00000368750.8
|
SPRR2E
|
small proline rich protein 2E |
| chr19_-_50968966 | 2.95 |
ENST00000376851.7
|
KLK6
|
kallikrein related peptidase 6 |
| chr19_-_50969567 | 2.87 |
ENST00000310157.7
|
KLK6
|
kallikrein related peptidase 6 |
| chr1_-_153057504 | 2.46 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chrX_+_70290077 | 1.96 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A |
| chr4_-_158173004 | 1.88 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr6_+_73696145 | 1.47 |
ENST00000287097.6
|
CD109
|
CD109 molecule |
| chr1_+_152908538 | 1.46 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr18_+_23949847 | 1.45 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr4_-_158173042 | 1.41 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr19_-_11339573 | 1.40 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr2_+_209580024 | 1.33 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr4_+_8229170 | 1.31 |
ENST00000511002.6
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr11_+_94768331 | 1.29 |
ENST00000317829.12
ENST00000433060.3 |
AMOTL1
|
angiomotin like 1 |
| chr11_-_48983826 | 1.28 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr5_+_69189536 | 1.19 |
ENST00000515001.5
ENST00000283006.7 ENST00000502689.1 |
CENPH
|
centromere protein H |
| chrX_+_65667645 | 1.18 |
ENST00000360270.7
|
MSN
|
moesin |
| chr6_-_111793871 | 1.15 |
ENST00000368667.6
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr9_+_5450503 | 1.08 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr4_+_143433491 | 1.05 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chr19_+_18386150 | 0.96 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr11_+_60396435 | 0.95 |
ENST00000395005.6
|
MS4A14
|
membrane spanning 4-domains A14 |
| chr11_+_61228377 | 0.92 |
ENST00000537932.5
|
PGA4
|
pepsinogen A4 |
| chr2_+_108377947 | 0.92 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr2_+_209579598 | 0.89 |
ENST00000445941.5
ENST00000673860.1 |
MAP2
|
microtubule associated protein 2 |
| chr2_+_190927649 | 0.89 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr2_+_157257687 | 0.89 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr14_-_55191534 | 0.87 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr5_-_11588842 | 0.80 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2 |
| chr9_+_107306459 | 0.79 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr2_+_17539964 | 0.78 |
ENST00000457525.5
|
VSNL1
|
visinin like 1 |
| chr11_-_101583985 | 0.78 |
ENST00000344327.8
|
TRPC6
|
transient receptor potential cation channel subfamily C member 6 |
| chr18_+_24155938 | 0.77 |
ENST00000582229.1
|
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr22_+_35717872 | 0.74 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L5 |
| chrX_+_2752024 | 0.70 |
ENST00000644266.2
ENST00000419513.7 ENST00000509484.3 ENST00000381174.10 |
XG
|
Xg glycoprotein (Xg blood group) |
| chr22_-_28306645 | 0.68 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr12_-_7936177 | 0.67 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr19_-_51723968 | 0.66 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr17_-_82840010 | 0.65 |
ENST00000269394.4
ENST00000572562.1 |
ZNF750
|
zinc finger protein 750 |
| chr3_+_42856021 | 0.63 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr18_-_31760864 | 0.63 |
ENST00000269205.7
ENST00000672005.1 |
SLC25A52
|
solute carrier family 25 member 52 |
| chr2_-_187513641 | 0.63 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr7_+_142332182 | 0.59 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr6_+_54083423 | 0.58 |
ENST00000460844.6
ENST00000370876.6 |
MLIP
|
muscular LMNA interacting protein |
| chr11_+_65638085 | 0.58 |
ENST00000534313.6
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr2_+_230759918 | 0.58 |
ENST00000614925.1
|
CAB39
|
calcium binding protein 39 |
| chr1_+_15152522 | 0.58 |
ENST00000428417.5
|
TMEM51
|
transmembrane protein 51 |
| chr14_-_106875069 | 0.57 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr17_+_7307961 | 0.57 |
ENST00000419711.6
ENST00000571955.5 ENST00000573714.5 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_-_11589019 | 0.57 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr1_-_197067234 | 0.56 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr11_-_111871271 | 0.56 |
ENST00000398006.6
|
ALG9
|
ALG9 alpha-1,2-mannosyltransferase |
| chr1_+_169795022 | 0.55 |
ENST00000359326.9
ENST00000496973.5 |
C1orf112
|
chromosome 1 open reading frame 112 |
| chr11_+_60396451 | 0.55 |
ENST00000300187.11
ENST00000526375.5 ENST00000531783.5 |
MS4A14
|
membrane spanning 4-domains A14 |
| chr5_+_136160986 | 0.55 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr6_+_69232406 | 0.55 |
ENST00000238918.12
|
ADGRB3
|
adhesion G protein-coupled receptor B3 |
| chr4_-_41214602 | 0.55 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr1_+_15152558 | 0.55 |
ENST00000376014.7
ENST00000451326.6 |
TMEM51
|
transmembrane protein 51 |
| chr4_+_70734419 | 0.54 |
ENST00000502653.5
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr16_+_14632906 | 0.54 |
ENST00000563971.5
ENST00000562442.5 ENST00000261658.7 |
BFAR
|
bifunctional apoptosis regulator |
| chr22_+_43923755 | 0.54 |
ENST00000423180.2
ENST00000216180.8 |
PNPLA3
|
patatin like phospholipase domain containing 3 |
| chr5_-_138338325 | 0.53 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr1_+_50103903 | 0.52 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr15_-_89211803 | 0.52 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr14_-_23365149 | 0.51 |
ENST00000216733.8
|
EFS
|
embryonal Fyn-associated substrate |
| chr7_+_1688119 | 0.50 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr5_+_115841878 | 0.50 |
ENST00000316788.12
|
AP3S1
|
adaptor related protein complex 3 subunit sigma 1 |
| chrX_+_108045050 | 0.48 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr3_-_197226351 | 0.48 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr17_+_7308339 | 0.47 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr6_+_57317602 | 0.47 |
ENST00000274891.10
ENST00000671770.1 ENST00000672107.1 ENST00000615550.5 |
PRIM2
|
DNA primase subunit 2 |
| chr16_-_20691256 | 0.46 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr17_+_68035636 | 0.46 |
ENST00000676857.1
ENST00000676594.1 ENST00000678388.1 |
KPNA2
|
karyopherin subunit alpha 2 |
| chr15_+_88635626 | 0.46 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr1_-_198540674 | 0.46 |
ENST00000489986.1
ENST00000367382.6 |
ATP6V1G3
|
ATPase H+ transporting V1 subunit G3 |
| chr1_+_86993009 | 0.45 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein |
| chr3_-_197184131 | 0.45 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_108378176 | 0.45 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr7_+_111091006 | 0.45 |
ENST00000451085.5
ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr2_+_201129483 | 0.44 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr14_-_106038355 | 0.43 |
ENST00000390597.3
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr17_-_49848017 | 0.43 |
ENST00000326219.5
ENST00000334568.8 ENST00000352793.6 ENST00000398154.5 ENST00000436235.5 |
TAC4
|
tachykinin precursor 4 |
| chr8_+_69466617 | 0.43 |
ENST00000525061.5
ENST00000260128.8 ENST00000458141.6 |
SULF1
|
sulfatase 1 |
| chr15_+_67067780 | 0.43 |
ENST00000679624.1
|
SMAD3
|
SMAD family member 3 |
| chr7_+_111091119 | 0.43 |
ENST00000308478.10
|
LRRN3
|
leucine rich repeat neuronal 3 |
| chr2_+_201129826 | 0.42 |
ENST00000457277.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrX_+_108044967 | 0.42 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrX_+_71578435 | 0.42 |
ENST00000373696.8
|
GCNA
|
germ cell nuclear acidic peptidase |
| chr4_+_118034480 | 0.40 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr11_+_55883297 | 0.40 |
ENST00000449290.6
|
TRIM51
|
tripartite motif-containing 51 |
| chr16_+_11965234 | 0.40 |
ENST00000562385.1
|
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr2_-_162152239 | 0.39 |
ENST00000418842.7
|
GCG
|
glucagon |
| chr1_+_153778178 | 0.39 |
ENST00000532853.5
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr3_-_197183849 | 0.39 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr11_-_89063631 | 0.39 |
ENST00000455756.6
|
GRM5
|
glutamate metabotropic receptor 5 |
| chr5_-_172006567 | 0.38 |
ENST00000517395.6
ENST00000265094.9 ENST00000393802.6 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr3_-_179266971 | 0.38 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr20_-_31406207 | 0.38 |
ENST00000376314.3
|
DEFB121
|
defensin beta 121 |
| chr1_+_206635573 | 0.37 |
ENST00000367108.7
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr11_-_107712049 | 0.36 |
ENST00000305991.3
|
SLN
|
sarcolipin |
| chrX_-_13734575 | 0.36 |
ENST00000519885.5
ENST00000458511.7 ENST00000380579.6 ENST00000683983.1 ENST00000683569.1 ENST00000359680.9 |
TRAPPC2
|
trafficking protein particle complex 2 |
| chr16_-_370514 | 0.36 |
ENST00000199706.13
|
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr12_-_102478539 | 0.35 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr12_-_11134644 | 0.35 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr1_+_206635521 | 0.35 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr6_+_31586269 | 0.34 |
ENST00000438075.7
|
LST1
|
leukocyte specific transcript 1 |
| chr1_+_6034980 | 0.34 |
ENST00000378092.6
ENST00000472700.7 |
KCNAB2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr16_+_31108294 | 0.33 |
ENST00000287507.7
ENST00000394950.7 ENST00000219794.11 ENST00000561755.1 |
BCKDK
|
branched chain keto acid dehydrogenase kinase |
| chr6_+_72366730 | 0.33 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr16_-_370338 | 0.33 |
ENST00000450882.1
ENST00000441883.5 ENST00000447696.5 ENST00000389675.6 |
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr2_+_201129318 | 0.33 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr2_-_162243375 | 0.32 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr5_-_151080978 | 0.32 |
ENST00000520931.5
ENST00000521591.6 ENST00000520695.5 ENST00000610535.5 ENST00000518977.5 ENST00000389378.6 ENST00000610874.4 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr10_+_5094405 | 0.31 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr14_-_106025628 | 0.31 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr16_+_11965193 | 0.31 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr3_-_98522514 | 0.31 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr5_-_172006817 | 0.31 |
ENST00000296933.10
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr3_-_197183806 | 0.30 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr6_+_31586124 | 0.30 |
ENST00000418507.6
ENST00000376096.5 ENST00000376099.5 ENST00000376110.7 |
LST1
|
leukocyte specific transcript 1 |
| chr3_-_197183963 | 0.30 |
ENST00000653795.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr5_+_108924585 | 0.30 |
ENST00000618353.1
|
FER
|
FER tyrosine kinase |
| chr17_-_676348 | 0.29 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr14_-_60165293 | 0.29 |
ENST00000554101.5
ENST00000557137.1 |
DHRS7
|
dehydrogenase/reductase 7 |
| chr22_+_24603147 | 0.29 |
ENST00000412658.5
ENST00000445029.5 ENST00000400382.6 ENST00000419133.5 ENST00000438643.6 ENST00000452551.5 ENST00000412898.5 ENST00000400380.5 ENST00000455483.5 ENST00000430289.5 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_45039509 | 0.29 |
ENST00000544944.6
|
CLASRP
|
CLK4 associating serine/arginine rich protein |
| chr10_+_69278492 | 0.29 |
ENST00000643399.2
|
HK1
|
hexokinase 1 |
| chr17_-_60392333 | 0.29 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr1_+_20290869 | 0.28 |
ENST00000289815.13
ENST00000375079.6 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr14_+_22207502 | 0.28 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr6_+_11093753 | 0.28 |
ENST00000416247.4
|
SMIM13
|
small integral membrane protein 13 |
| chr4_+_108650585 | 0.27 |
ENST00000613215.4
ENST00000361564.9 |
OSTC
|
oligosaccharyltransferase complex non-catalytic subunit |
| chr3_-_108058361 | 0.27 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr7_-_27165517 | 0.27 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr7_+_114922854 | 0.27 |
ENST00000423503.1
ENST00000427207.5 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr2_-_97094882 | 0.27 |
ENST00000414820.6
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr2_+_89947508 | 0.26 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr3_-_33659097 | 0.26 |
ENST00000461133.8
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_60056587 | 0.25 |
ENST00000395032.6
ENST00000358152.6 |
MS4A3
|
membrane spanning 4-domains A3 |
| chr7_-_48029102 | 0.25 |
ENST00000297325.9
ENST00000412142.5 ENST00000395572.6 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr3_-_42875871 | 0.25 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr14_-_106803221 | 0.25 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr7_-_4959172 | 0.25 |
ENST00000401401.8
ENST00000406755.5 ENST00000404774.7 ENST00000612910.1 |
MMD2
|
monocyte to macrophage differentiation associated 2 |
| chr1_+_56854764 | 0.24 |
ENST00000361249.4
|
C8A
|
complement C8 alpha chain |
| chr14_+_24070837 | 0.24 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr2_+_218245426 | 0.24 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex subunit 2 |
| chr4_-_170027209 | 0.24 |
ENST00000393702.7
|
MFAP3L
|
microfibril associated protein 3 like |
| chr1_+_196774813 | 0.24 |
ENST00000471440.6
ENST00000391985.7 ENST00000617219.1 ENST00000367425.9 |
CFHR3
|
complement factor H related 3 |
| chr19_-_1021114 | 0.23 |
ENST00000333175.9
ENST00000356663.8 |
TMEM259
|
transmembrane protein 259 |
| chr17_-_41382298 | 0.23 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr14_+_94581407 | 0.23 |
ENST00000553511.1
ENST00000329597.12 ENST00000554633.5 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin family A member 5 |
| chr7_+_4682252 | 0.23 |
ENST00000328914.5
|
FOXK1
|
forkhead box K1 |
| chr17_-_49209367 | 0.23 |
ENST00000300406.6
ENST00000511277.5 ENST00000511673.1 |
GNGT2
|
G protein subunit gamma transducin 2 |
| chr2_-_182242031 | 0.23 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr12_-_122966438 | 0.22 |
ENST00000344275.11
ENST00000442833.6 ENST00000280560.13 ENST00000540285.5 ENST00000346530.9 |
ABCB9
|
ATP binding cassette subfamily B member 9 |
| chr10_+_134703 | 0.22 |
ENST00000509513.6
|
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr14_+_94581388 | 0.22 |
ENST00000554866.5
ENST00000556775.5 |
SERPINA5
|
serpin family A member 5 |
| chr6_-_2744126 | 0.22 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr12_+_9971402 | 0.22 |
ENST00000304361.9
ENST00000396507.7 ENST00000434319.6 |
CLEC12A
|
C-type lectin domain family 12 member A |
| chr1_+_159439722 | 0.22 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr15_+_75647877 | 0.22 |
ENST00000308527.6
|
SNX33
|
sorting nexin 33 |
| chr12_-_52703522 | 0.21 |
ENST00000341809.8
|
KRT77
|
keratin 77 |
| chr5_-_115841548 | 0.21 |
ENST00000509910.2
ENST00000500945.2 |
ATG12
|
autophagy related 12 |
| chr3_-_126357399 | 0.21 |
ENST00000296233.4
|
KLF15
|
Kruppel like factor 15 |
| chr3_-_195583931 | 0.21 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_+_151733919 | 0.21 |
ENST00000356517.4
|
AADACL2
|
arylacetamide deacetylase like 2 |
| chr19_+_38899680 | 0.21 |
ENST00000576510.5
ENST00000392079.7 |
NFKBIB
|
NFKB inhibitor beta |
| chrX_-_70289888 | 0.21 |
ENST00000239666.9
ENST00000374454.1 |
PDZD11
|
PDZ domain containing 11 |
| chr17_+_47253817 | 0.21 |
ENST00000559488.7
ENST00000571680.1 |
ITGB3
|
integrin subunit beta 3 |
| chr16_+_1966820 | 0.21 |
ENST00000569210.6
ENST00000569714.6 |
RNF151
|
ring finger protein 151 |
| chr8_+_21966215 | 0.20 |
ENST00000433566.8
|
XPO7
|
exportin 7 |
| chr9_-_92482499 | 0.20 |
ENST00000375544.7
|
ASPN
|
asporin |
| chr2_-_32265732 | 0.20 |
ENST00000360906.9
ENST00000342905.10 |
NLRC4
|
NLR family CARD domain containing 4 |
| chr9_-_21368962 | 0.20 |
ENST00000610660.1
|
IFNA13
|
interferon alpha 13 |
| chr12_-_52406335 | 0.20 |
ENST00000257974.3
|
KRT82
|
keratin 82 |
| chr2_-_39779217 | 0.20 |
ENST00000505747.6
|
THUMPD2
|
THUMP domain containing 2 |
| chr6_-_56542802 | 0.19 |
ENST00000524186.1
|
DST
|
dystonin |
| chr14_+_76826372 | 0.19 |
ENST00000393774.7
ENST00000555189.1 |
LRRC74A
|
leucine rich repeat containing 74A |
| chr3_-_38816217 | 0.19 |
ENST00000449082.3
ENST00000655275.1 |
SCN10A
|
sodium voltage-gated channel alpha subunit 10 |
| chr8_-_94436926 | 0.19 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein |
| chr7_-_88795732 | 0.19 |
ENST00000297203.3
|
TEX47
|
testis expressed 47 |
| chr4_+_87799546 | 0.19 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chrX_-_103502853 | 0.18 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr12_+_4590075 | 0.18 |
ENST00000540757.6
|
DYRK4
|
dual specificity tyrosine phosphorylation regulated kinase 4 |
| chr2_+_129979641 | 0.18 |
ENST00000410061.4
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr20_+_35968566 | 0.17 |
ENST00000373973.7
ENST00000349339.5 ENST00000489667.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr11_-_26722051 | 0.17 |
ENST00000396005.8
|
SLC5A12
|
solute carrier family 5 member 12 |
| chr4_+_69931066 | 0.17 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr3_+_184300564 | 0.17 |
ENST00000435761.5
ENST00000439383.5 |
PSMD2
|
proteasome 26S subunit, non-ATPase 2 |
| chr9_-_21217311 | 0.17 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr2_-_227717981 | 0.17 |
ENST00000409456.2
ENST00000409287.5 ENST00000644224.2 ENST00000456524.6 |
SLC19A3
|
solute carrier family 19 member 3 |
| chr15_+_90875688 | 0.16 |
ENST00000681865.1
|
FURIN
|
furin, paired basic amino acid cleaving enzyme |
| chr6_-_31958935 | 0.16 |
ENST00000441998.5
ENST00000444811.6 ENST00000375429.8 |
NELFE
|
negative elongation factor complex member E |
| chr9_-_124415421 | 0.16 |
ENST00000259457.8
ENST00000441097.1 |
PSMB7
|
proteasome 20S subunit beta 7 |
| chr19_-_20039492 | 0.15 |
ENST00000397165.7
|
ZNF682
|
zinc finger protein 682 |
| chr16_-_18900044 | 0.15 |
ENST00000565224.5
ENST00000330588.4 |
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr3_+_119579577 | 0.15 |
ENST00000478927.5
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr2_+_171687501 | 0.15 |
ENST00000508530.5
|
DYNC1I2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr10_+_27532521 | 0.15 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 1.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 5.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 1.5 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 1.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.7 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.6 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 1.9 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 2.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0016488 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 5.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.3 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.5 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 0.3 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.4 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 1.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 1.3 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 2.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 1.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 0.8 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.2 | 0.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 0.5 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 6.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.9 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 2.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 2.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.7 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.2 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 10.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.3 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.2 | GO:0023029 | MHC class Ib protein binding(GO:0023029) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 3.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |