|
chr17_-_40703744
Show fit
|
7.87 |
ENST00000264651.3
|
KRT24
|
keratin 24
|
|
chr8_+_53851786
Show fit
|
2.47 |
ENST00000297313.8
ENST00000344277.10
|
RGS20
|
regulator of G protein signaling 20
|
|
chr19_-_51034840
Show fit
|
2.45 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12
|
|
chr19_-_51034892
Show fit
|
2.43 |
ENST00000319590.8
ENST00000250351.4
|
KLK12
|
kallikrein related peptidase 12
|
|
chr19_-_51034993
Show fit
|
2.26 |
ENST00000684732.1
|
KLK12
|
kallikrein related peptidase 12
|
|
chr15_-_74202742
Show fit
|
1.54 |
ENST00000395105.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6
|
|
chr5_-_150289941
Show fit
|
1.44 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr16_+_57976435
Show fit
|
1.21 |
ENST00000290871.10
ENST00000441824.4
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr5_-_150289764
Show fit
|
1.13 |
ENST00000671881.1
ENST00000672752.1
ENST00000510347.2
ENST00000672829.1
ENST00000348628.11
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr1_+_180928133
Show fit
|
1.12 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614
|
|
chr5_-_150289625
Show fit
|
1.10 |
ENST00000683332.1
ENST00000398376.8
ENST00000672785.1
ENST00000672396.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr15_+_33310946
Show fit
|
1.07 |
ENST00000415757.7
ENST00000634891.2
ENST00000389232.9
ENST00000622037.1
|
RYR3
|
ryanodine receptor 3
|
|
chr2_-_65432591
Show fit
|
1.06 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2
|
|
chr10_+_123135938
Show fit
|
0.98 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3
|
|
chr12_+_6840800
Show fit
|
0.97 |
ENST00000541978.5
ENST00000229264.8
ENST00000435982.6
|
GNB3
|
G protein subunit beta 3
|
|
chr3_-_149377637
Show fit
|
0.93 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr6_+_130018565
Show fit
|
0.89 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr8_-_90082871
Show fit
|
0.88 |
ENST00000265431.7
|
CALB1
|
calbindin 1
|
|
chr11_-_62984690
Show fit
|
0.86 |
ENST00000421062.2
ENST00000458333.6
|
SLC22A6
|
solute carrier family 22 member 6
|
|
chrX_+_70290077
Show fit
|
0.85 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A
|
|
chr18_-_31102411
Show fit
|
0.77 |
ENST00000251081.8
ENST00000280904.11
ENST00000682357.1
ENST00000648081.1
|
DSC2
|
desmocollin 2
|
|
chr14_-_94770102
Show fit
|
0.72 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox
|
|
chrX_+_70268324
Show fit
|
0.70 |
ENST00000307959.9
ENST00000620997.4
ENST00000480877.6
|
ARR3
|
arrestin 3
|
|
chr9_+_113536497
Show fit
|
0.69 |
ENST00000462143.5
|
RGS3
|
regulator of G protein signaling 3
|
|
chr19_-_51034727
Show fit
|
0.67 |
ENST00000525263.5
|
KLK12
|
kallikrein related peptidase 12
|
|
chr5_-_150290093
Show fit
|
0.67 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chrX_+_70268305
Show fit
|
0.63 |
ENST00000374495.7
|
ARR3
|
arrestin 3
|
|
chrX_+_136197020
Show fit
|
0.60 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1
|
|
chr10_-_84241538
Show fit
|
0.59 |
ENST00000372105.4
|
LRIT1
|
leucine rich repeat, Ig-like and transmembrane domains 1
|
|
chr2_+_218607861
Show fit
|
0.58 |
ENST00000450993.7
|
PLCD4
|
phospholipase C delta 4
|
|
chr17_-_55511434
Show fit
|
0.57 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36
|
|
chr2_-_70553440
Show fit
|
0.56 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha
|
|
chr10_-_104085847
Show fit
|
0.55 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain
|
|
chr6_-_35512863
Show fit
|
0.55 |
ENST00000428978.1
ENST00000614066.4
ENST00000322263.8
|
TULP1
|
TUB like protein 1
|
|
chr1_+_55039511
Show fit
|
0.55 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr16_-_57971086
Show fit
|
0.54 |
ENST00000564448.5
ENST00000311183.8
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1
|
|
chr5_-_115626161
Show fit
|
0.54 |
ENST00000282382.8
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough
|
|
chrX_-_124963768
Show fit
|
0.50 |
ENST00000371130.7
ENST00000422452.2
|
TENM1
|
teneurin transmembrane protein 1
|
|
chr6_+_35342535
Show fit
|
0.49 |
ENST00000360694.8
ENST00000418635.6
ENST00000448077.6
|
PPARD
|
peroxisome proliferator activated receptor delta
|
|
chr6_+_35342614
Show fit
|
0.49 |
ENST00000337400.6
ENST00000311565.4
|
PPARD
|
peroxisome proliferator activated receptor delta
|
|
chr12_-_52796110
Show fit
|
0.48 |
ENST00000417996.2
|
KRT3
|
keratin 3
|
|
chr12_-_52777343
Show fit
|
0.47 |
ENST00000332411.2
|
KRT76
|
keratin 76
|
|
chr3_+_50611871
Show fit
|
0.47 |
ENST00000446044.5
|
MAPKAPK3
|
MAPK activated protein kinase 3
|
|
chrX_+_136197039
Show fit
|
0.45 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1
|
|
chrX_+_136196750
Show fit
|
0.44 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_+_130931893
Show fit
|
0.44 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chr2_-_70553638
Show fit
|
0.44 |
ENST00000444975.5
ENST00000445399.5
ENST00000295400.11
ENST00000418333.6
|
TGFA
|
transforming growth factor alpha
|
|
chr3_-_48188356
Show fit
|
0.43 |
ENST00000351231.7
ENST00000437972.1
ENST00000302506.8
|
CDC25A
|
cell division cycle 25A
|
|
chr11_-_111912871
Show fit
|
0.43 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B
|
|
chr6_+_106087580
Show fit
|
0.42 |
ENST00000424894.1
ENST00000648754.1
|
PRDM1
|
PR/SET domain 1
|
|
chr17_-_45132505
Show fit
|
0.41 |
ENST00000619929.5
|
PLCD3
|
phospholipase C delta 3
|
|
chr12_-_1918473
Show fit
|
0.40 |
ENST00000586184.5
ENST00000587995.5
ENST00000585732.1
|
CACNA2D4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4
|
|
chr8_-_132111159
Show fit
|
0.40 |
ENST00000673615.1
ENST00000434736.6
|
HHLA1
|
HERV-H LTR-associating 1
|
|
chr15_-_34318761
Show fit
|
0.39 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6
|
|
chr8_-_25424260
Show fit
|
0.39 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1
|
|
chr6_-_138572523
Show fit
|
0.38 |
ENST00000427025.6
|
NHSL1
|
NHS like 1
|
|
chr11_-_62984957
Show fit
|
0.38 |
ENST00000377871.7
ENST00000360421.9
|
SLC22A6
|
solute carrier family 22 member 6
|
|
chr20_+_44355692
Show fit
|
0.38 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr12_+_53299682
Show fit
|
0.37 |
ENST00000267103.10
ENST00000548632.5
|
MYG1
|
MYG1 exonuclease
|
|
chr7_-_128775793
Show fit
|
0.37 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive
|
|
chr17_-_62808339
Show fit
|
0.37 |
ENST00000583600.5
|
MARCHF10
|
membrane associated ring-CH-type finger 10
|
|
chr14_-_63318933
Show fit
|
0.37 |
ENST00000621500.2
|
GPHB5
|
glycoprotein hormone subunit beta 5
|
|
chr12_+_53300027
Show fit
|
0.36 |
ENST00000549488.5
|
MYG1
|
MYG1 exonuclease
|
|
chr17_-_62808299
Show fit
|
0.36 |
ENST00000311269.10
|
MARCHF10
|
membrane associated ring-CH-type finger 10
|
|
chr6_+_31587268
Show fit
|
0.36 |
ENST00000396101.7
ENST00000490742.5
|
LST1
|
leukocyte specific transcript 1
|
|
chr1_-_150235972
Show fit
|
0.35 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr1_+_11691688
Show fit
|
0.35 |
ENST00000294485.6
|
DRAXIN
|
dorsal inhibitory axon guidance protein
|
|
chr20_-_50115935
Show fit
|
0.33 |
ENST00000340309.7
ENST00000415862.6
ENST00000371677.7
|
UBE2V1
|
ubiquitin conjugating enzyme E2 V1
|
|
chr15_+_58138368
Show fit
|
0.32 |
ENST00000219919.9
ENST00000536493.1
|
AQP9
|
aquaporin 9
|
|
chr1_+_159439722
Show fit
|
0.32 |
ENST00000641630.1
ENST00000423932.6
|
OR10J1
|
olfactory receptor family 10 subfamily J member 1
|
|
chr9_+_18474206
Show fit
|
0.32 |
ENST00000276935.6
|
ADAMTSL1
|
ADAMTS like 1
|
|
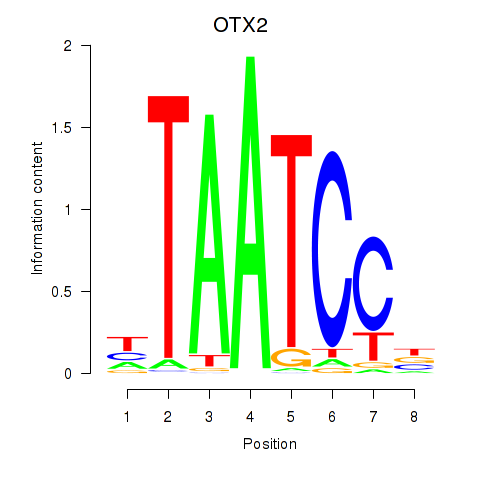
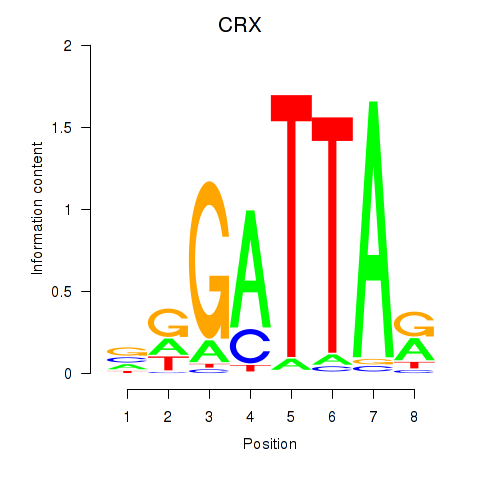
chr19_+_47821907
Show fit
|
0.32 |
ENST00000539067.5
ENST00000221996.12
ENST00000613299.1
|
CRX
|
cone-rod homeobox
|
|
chr12_+_77830886
Show fit
|
0.31 |
ENST00000397909.7
ENST00000549464.5
|
NAV3
|
neuron navigator 3
|
|
chr3_+_111674654
Show fit
|
0.31 |
ENST00000636933.1
ENST00000393934.7
ENST00000477665.2
|
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2
|
|
chr4_-_152679984
Show fit
|
0.31 |
ENST00000304385.8
ENST00000504064.1
|
TMEM154
|
transmembrane protein 154
|
|
chr1_-_99766620
Show fit
|
0.30 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1
|
|
chr10_+_47348351
Show fit
|
0.30 |
ENST00000584701.2
|
RBP3
|
retinol binding protein 3
|
|
chr8_+_69466617
Show fit
|
0.29 |
ENST00000525061.5
ENST00000260128.8
ENST00000458141.6
|
SULF1
|
sulfatase 1
|
|
chr11_+_73264479
Show fit
|
0.29 |
ENST00000393592.7
ENST00000540342.6
ENST00000679753.1
ENST00000542092.5
ENST00000349767.6
|
P2RY6
|
pyrimidinergic receptor P2Y6
|
|
chr18_-_66604076
Show fit
|
0.29 |
ENST00000540086.5
ENST00000580157.2
ENST00000262150.7
|
CDH19
|
cadherin 19
|
|
chr6_+_34466059
Show fit
|
0.28 |
ENST00000620693.4
ENST00000244458.7
ENST00000374043.6
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr16_-_57971121
Show fit
|
0.28 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1
|
|
chr6_-_30690968
Show fit
|
0.28 |
ENST00000376420.9
ENST00000376421.7
|
NRM
|
nurim
|
|
chr9_+_34179005
Show fit
|
0.28 |
ENST00000625521.2
ENST00000379186.8
ENST00000297661.9
ENST00000626262.2
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr6_-_65707214
Show fit
|
0.28 |
ENST00000370621.7
ENST00000393380.6
ENST00000503581.6
|
EYS
|
eyes shut homolog
|
|
chr5_-_142325001
Show fit
|
0.28 |
ENST00000344120.4
ENST00000434127.3
|
SPRY4
|
sprouty RTK signaling antagonist 4
|
|
chr1_+_18631006
Show fit
|
0.28 |
ENST00000375375.7
|
PAX7
|
paired box 7
|
|
chr5_-_96433214
Show fit
|
0.28 |
ENST00000311106.8
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr19_+_45498439
Show fit
|
0.28 |
ENST00000451287.7
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative)
|
|
chr11_+_124865425
Show fit
|
0.27 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3
|
|
chr17_-_73644435
Show fit
|
0.27 |
ENST00000392650.8
|
SDK2
|
sidekick cell adhesion molecule 2
|
|
chr17_+_45241067
Show fit
|
0.26 |
ENST00000587489.5
|
FMNL1
|
formin like 1
|
|
chr5_-_88883199
Show fit
|
0.26 |
ENST00000514015.5
ENST00000503075.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_-_48044037
Show fit
|
0.26 |
ENST00000293255.3
|
CABP5
|
calcium binding protein 5
|
|
chr3_-_54928044
Show fit
|
0.26 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1
|
|
chr4_-_101347327
Show fit
|
0.25 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr15_+_92900338
Show fit
|
0.25 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_+_99606833
Show fit
|
0.25 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13
|
|
chr3_-_50611767
Show fit
|
0.25 |
ENST00000443053.6
ENST00000348721.4
|
CISH
|
cytokine inducible SH2 containing protein
|
|
chr17_-_41505597
Show fit
|
0.25 |
ENST00000336861.7
ENST00000246635.8
ENST00000587544.5
ENST00000587435.1
|
KRT13
|
keratin 13
|
|
chrX_-_21658324
Show fit
|
0.24 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34
|
|
chr9_+_100442271
Show fit
|
0.24 |
ENST00000502978.1
|
MSANTD3-TMEFF1
|
MSANTD3-TMEFF1 readthrough
|
|
chr7_+_28412511
Show fit
|
0.24 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_+_69145463
Show fit
|
0.24 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2
|
|
chr2_-_29921580
Show fit
|
0.24 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase
|
|
chr11_-_111923722
Show fit
|
0.24 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B
|
|
chr4_-_101347492
Show fit
|
0.24 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr3_-_138132381
Show fit
|
0.24 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase
|
|
chrX_-_54357993
Show fit
|
0.23 |
ENST00000375169.7
ENST00000354646.6
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr11_+_20022550
Show fit
|
0.23 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2
|
|
chr1_-_150235995
Show fit
|
0.23 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chrX_+_80420466
Show fit
|
0.23 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D
|
|
chr2_+_191276885
Show fit
|
0.23 |
ENST00000392316.5
|
MYO1B
|
myosin IB
|
|
chr7_-_84194781
Show fit
|
0.23 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A
|
|
chr18_+_616672
Show fit
|
0.23 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1
|
|
chr4_-_101347471
Show fit
|
0.23 |
ENST00000323055.10
ENST00000512215.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr17_+_17042433
Show fit
|
0.22 |
ENST00000651222.2
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr18_+_616711
Show fit
|
0.22 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1
|
|
chr5_+_136058849
Show fit
|
0.22 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced
|
|
chr4_-_101346842
Show fit
|
0.21 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha
|
|
chr18_+_34710249
Show fit
|
0.21 |
ENST00000680346.1
ENST00000348997.9
ENST00000681274.1
ENST00000680822.1
ENST00000680767.2
ENST00000597599.5
ENST00000444659.6
|
DTNA
|
dystrobrevin alpha
|
|
chr1_-_150235943
Show fit
|
0.21 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr6_+_31587185
Show fit
|
0.21 |
ENST00000376092.7
ENST00000376086.7
ENST00000303757.12
ENST00000376093.6
|
LST1
|
leukocyte specific transcript 1
|
|
chr17_+_8002610
Show fit
|
0.21 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal
|
|
chr17_+_77281429
Show fit
|
0.21 |
ENST00000591198.5
ENST00000427177.6
|
SEPTIN9
|
septin 9
|
|
chr6_+_31587049
Show fit
|
0.21 |
ENST00000376089.6
ENST00000396112.6
|
LST1
|
leukocyte specific transcript 1
|
|
chr9_+_18474100
Show fit
|
0.20 |
ENST00000327883.11
ENST00000431052.6
ENST00000380570.8
ENST00000380548.9
|
ADAMTSL1
|
ADAMTS like 1
|
|
chrX_+_41447322
Show fit
|
0.20 |
ENST00000378220.2
ENST00000342595.2
|
NYX
|
nyctalopin
|
|
chr9_-_128724088
Show fit
|
0.20 |
ENST00000406904.2
ENST00000452105.5
ENST00000372667.9
ENST00000372663.9
|
ZDHHC12
|
zinc finger DHHC-type palmitoyltransferase 12
|
|
chr19_-_42427379
Show fit
|
0.20 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type
|
|
chr1_-_11691608
Show fit
|
0.20 |
ENST00000376667.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr12_-_110499546
Show fit
|
0.20 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component
|
|
chr9_-_92482350
Show fit
|
0.19 |
ENST00000375543.2
|
ASPN
|
asporin
|
|
chr18_-_37565825
Show fit
|
0.19 |
ENST00000603232.6
|
CELF4
|
CUGBP Elav-like family member 4
|
|
chr1_-_11691646
Show fit
|
0.19 |
ENST00000235310.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr3_-_197226351
Show fit
|
0.19 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1
|
|
chr15_-_65115185
Show fit
|
0.19 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like
|
|
chr5_-_36301883
Show fit
|
0.19 |
ENST00000502994.5
ENST00000515759.5
ENST00000296604.8
|
RANBP3L
|
RAN binding protein 3 like
|
|
chr10_-_75109085
Show fit
|
0.19 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr7_+_80602200
Show fit
|
0.19 |
ENST00000534394.5
|
CD36
|
CD36 molecule
|
|
chr17_-_48613468
Show fit
|
0.18 |
ENST00000498634.2
|
HOXB8
|
homeobox B8
|
|
chr7_+_80602150
Show fit
|
0.18 |
ENST00000309881.11
|
CD36
|
CD36 molecule
|
|
chr19_-_3772211
Show fit
|
0.18 |
ENST00000555978.5
ENST00000555633.3
|
RAX2
|
retina and anterior neural fold homeobox 2
|
|
chr3_+_4493340
Show fit
|
0.18 |
ENST00000357086.10
ENST00000354582.12
ENST00000649015.2
ENST00000467056.6
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr3_-_98522514
Show fit
|
0.17 |
ENST00000503004.5
ENST00000506575.1
ENST00000513452.5
ENST00000515620.5
|
CLDND1
|
claudin domain containing 1
|
|
chr9_-_92482499
Show fit
|
0.17 |
ENST00000375544.7
|
ASPN
|
asporin
|
|
chr1_-_11691471
Show fit
|
0.17 |
ENST00000376672.5
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr10_+_102226293
Show fit
|
0.17 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3
|
|
chr2_+_33436304
Show fit
|
0.17 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3
|
|
chr5_-_11589019
Show fit
|
0.16 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2
|
|
chr2_-_55010348
Show fit
|
0.16 |
ENST00000394609.6
|
RTN4
|
reticulon 4
|
|
chr16_+_81238682
Show fit
|
0.16 |
ENST00000258168.7
ENST00000564552.1
|
BCO1
|
beta-carotene oxygenase 1
|
|
chr10_-_75109172
Show fit
|
0.16 |
ENST00000372700.7
ENST00000473072.2
ENST00000491677.6
ENST00000372702.7
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr3_+_129528632
Show fit
|
0.16 |
ENST00000296271.4
|
RHO
|
rhodopsin
|
|
chr2_+_120013068
Show fit
|
0.15 |
ENST00000443902.6
ENST00000263713.10
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chr13_-_35476682
Show fit
|
0.15 |
ENST00000379919.6
|
MAB21L1
|
mab-21 like 1
|
|
chr2_-_61537740
Show fit
|
0.15 |
ENST00000678081.1
ENST00000676889.1
ENST00000677850.1
ENST00000676789.1
|
XPO1
|
exportin 1
|
|
chr8_-_80080816
Show fit
|
0.15 |
ENST00000520527.5
ENST00000517427.5
ENST00000379097.7
ENST00000448733.3
|
TPD52
|
tumor protein D52
|
|
chr10_+_24239181
Show fit
|
0.15 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217
|
|
chr11_-_11353241
Show fit
|
0.15 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3
|
|
chr3_+_191329020
Show fit
|
0.14 |
ENST00000392456.4
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr5_-_11588842
Show fit
|
0.14 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2
|
|
chr12_-_66803980
Show fit
|
0.14 |
ENST00000539540.5
ENST00000540433.5
ENST00000541947.1
ENST00000538373.1
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr17_-_48615261
Show fit
|
0.14 |
ENST00000239144.5
|
HOXB8
|
homeobox B8
|
|
chr15_+_92900189
Show fit
|
0.14 |
ENST00000626874.2
ENST00000627622.1
ENST00000629346.2
ENST00000628375.2
ENST00000420239.7
ENST00000394196.9
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr1_-_201112420
Show fit
|
0.14 |
ENST00000362061.4
ENST00000681874.1
|
CACNA1S
|
calcium voltage-gated channel subunit alpha1 S
|
|
chr7_-_126533850
Show fit
|
0.13 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8
|
|
chr14_-_34630109
Show fit
|
0.13 |
ENST00000396526.7
|
SNX6
|
sorting nexin 6
|
|
chr21_-_33643926
Show fit
|
0.13 |
ENST00000438788.1
|
CRYZL1
|
crystallin zeta like 1
|
|
chr1_-_150236064
Show fit
|
0.13 |
ENST00000532744.2
ENST00000369114.9
ENST00000369115.3
ENST00000583931.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr6_+_151809105
Show fit
|
0.13 |
ENST00000427531.6
|
ESR1
|
estrogen receptor 1
|
|
chr10_-_110304894
Show fit
|
0.13 |
ENST00000369603.10
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr5_-_88883420
Show fit
|
0.13 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr10_+_93612532
Show fit
|
0.13 |
ENST00000371447.4
|
PDE6C
|
phosphodiesterase 6C
|
|
chr3_-_170908626
Show fit
|
0.12 |
ENST00000295822.7
ENST00000474096.5
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr5_+_150660841
Show fit
|
0.12 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3
|
|
chr1_-_53142617
Show fit
|
0.12 |
ENST00000371491.4
ENST00000371494.9
|
SLC1A7
|
solute carrier family 1 member 7
|
|
chr1_-_151992571
Show fit
|
0.12 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10
|
|
chr19_+_39498938
Show fit
|
0.12 |
ENST00000356433.10
ENST00000596614.5
ENST00000205143.4
|
DLL3
|
delta like canonical Notch ligand 3
|
|
chr1_-_173205543
Show fit
|
0.11 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4
|
|
chr9_+_85941121
Show fit
|
0.11 |
ENST00000361671.10
|
NAA35
|
N-alpha-acetyltransferase 35, NatC auxiliary subunit
|
|
chr2_+_218607914
Show fit
|
0.11 |
ENST00000417849.5
|
PLCD4
|
phospholipase C delta 4
|
|
chr2_+_120013111
Show fit
|
0.11 |
ENST00000331393.8
ENST00000443124.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chr1_+_20070156
Show fit
|
0.11 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V
|
|
chr1_+_101237009
Show fit
|
0.11 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr10_+_100745711
Show fit
|
0.11 |
ENST00000370296.6
ENST00000428433.5
|
PAX2
|
paired box 2
|
|
chr10_+_50990864
Show fit
|
0.11 |
ENST00000401604.8
|
PRKG1
|
protein kinase cGMP-dependent 1
|
|
chr4_+_125314918
Show fit
|
0.11 |
ENST00000674496.2
ENST00000394329.9
|
FAT4
|
FAT atypical cadherin 4
|
|
chr5_-_41510623
Show fit
|
0.10 |
ENST00000328457.5
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3
|
|
chr10_+_102132994
Show fit
|
0.10 |
ENST00000413464.6
ENST00000278070.7
|
PPRC1
|
PPARG related coactivator 1
|
|
chr12_+_80707625
Show fit
|
0.10 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6
|
|
chrX_-_154097731
Show fit
|
0.10 |
ENST00000628176.2
|
MECP2
|
methyl-CpG binding protein 2
|
|
chr11_+_61755372
Show fit
|
0.10 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor
|
|
chr5_-_77776321
Show fit
|
0.10 |
ENST00000380377.9
|
TBCA
|
tubulin folding cofactor A
|
|
chr6_-_106975616
Show fit
|
0.10 |
ENST00000610952.1
|
CD24
|
CD24 molecule
|
|
chr8_-_103501890
Show fit
|
0.09 |
ENST00000649416.1
|
ENSG00000285982.1
|
novel protein
|
|
chr8_+_64580357
Show fit
|
0.09 |
ENST00000321870.3
|
BHLHE22
|
basic helix-loop-helix family member e22
|
|
chr12_-_86256299
Show fit
|
0.09 |
ENST00000552808.6
ENST00000547225.5
|
MGAT4C
|
MGAT4 family member C
|
|
chr12_-_118190510
Show fit
|
0.09 |
ENST00000540561.5
ENST00000537952.1
ENST00000537822.1
|
TAOK3
|
TAO kinase 3
|
|
chr9_-_28670285
Show fit
|
0.09 |
ENST00000379992.6
ENST00000308675.5
ENST00000613945.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|