Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PAX6
Z-value: 0.52
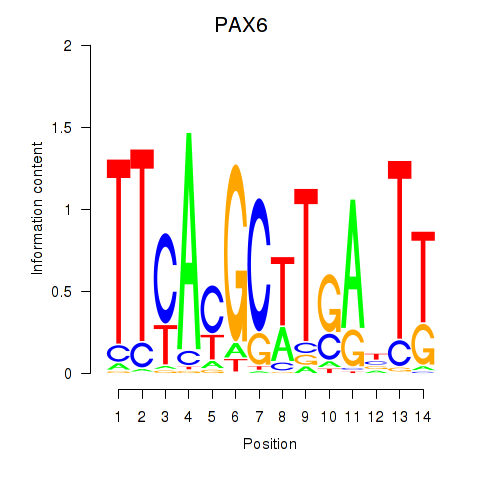
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.24 | PAX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg38_v1_chr11_-_31812171_31812342 | 0.24 | 2.0e-01 | Click! |
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_35528221 | 0.55 |
ENST00000588674.5
ENST00000452271.7 ENST00000518157.1 |
SBSN
|
suprabasin |
| chrX_-_66040107 | 0.54 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chrX_-_66040072 | 0.53 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_54623518 | 0.51 |
ENST00000302250.7
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151 member A |
| chr11_-_58575846 | 0.48 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr14_+_88005128 | 0.48 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65 |
| chr4_-_173399102 | 0.47 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr8_+_66043413 | 0.43 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr3_+_141262614 | 0.41 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr12_-_69699285 | 0.38 |
ENST00000553096.5
ENST00000330891.10 |
BEST3
|
bestrophin 3 |
| chr12_+_10307950 | 0.37 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr6_-_42194947 | 0.37 |
ENST00000230361.4
|
GUCA1B
|
guanylate cyclase activator 1B |
| chr19_-_51630401 | 0.33 |
ENST00000683636.1
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr5_+_96876480 | 0.33 |
ENST00000437043.8
ENST00000379904.8 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr22_-_17221841 | 0.32 |
ENST00000449907.7
ENST00000441548.1 ENST00000399839.5 |
ADA2
|
adenosine deaminase 2 |
| chr4_-_10684749 | 0.31 |
ENST00000226951.11
|
CLNK
|
cytokine dependent hematopoietic cell linker |
| chr3_+_87792697 | 0.30 |
ENST00000319595.7
|
HTR1F
|
5-hydroxytryptamine receptor 1F |
| chr7_+_143222037 | 0.29 |
ENST00000408947.4
|
TAS2R40
|
taste 2 receptor member 40 |
| chr10_-_91909476 | 0.29 |
ENST00000311575.6
|
FGFBP3
|
fibroblast growth factor binding protein 3 |
| chr11_-_102530738 | 0.29 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr4_-_67883987 | 0.27 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr12_+_56521990 | 0.26 |
ENST00000550726.5
ENST00000542360.1 |
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr15_-_28174423 | 0.26 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr3_-_195583931 | 0.25 |
ENST00000343267.8
ENST00000421243.5 ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr1_+_147902789 | 0.25 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr12_-_121039204 | 0.25 |
ENST00000620239.5
|
OASL
|
2'-5'-oligoadenylate synthetase like |
| chr7_-_80922354 | 0.23 |
ENST00000419255.6
|
SEMA3C
|
semaphorin 3C |
| chr2_+_90021567 | 0.23 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr12_+_56521951 | 0.23 |
ENST00000552247.6
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr20_-_45515612 | 0.23 |
ENST00000217428.7
|
SPINT3
|
serine peptidase inhibitor, Kunitz type 3 |
| chr12_-_84912816 | 0.23 |
ENST00000680469.1
ENST00000450363.4 ENST00000681106.1 |
SLC6A15
|
solute carrier family 6 member 15 |
| chr9_-_16870702 | 0.23 |
ENST00000380667.6
ENST00000545497.5 ENST00000486514.5 |
BNC2
|
basonuclin 2 |
| chr7_-_97872394 | 0.22 |
ENST00000455086.5
ENST00000394308.8 ENST00000453600.5 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr18_+_13611910 | 0.21 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr10_+_89332484 | 0.21 |
ENST00000371811.4
ENST00000680037.1 ENST00000679583.1 ENST00000679897.1 |
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3 |
| chr6_+_25652272 | 0.20 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr5_+_322570 | 0.20 |
ENST00000510400.5
ENST00000316418.10 |
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr19_+_35329161 | 0.20 |
ENST00000341773.10
ENST00000600131.5 ENST00000595780.5 ENST00000597916.5 ENST00000593867.5 ENST00000600424.5 ENST00000599811.5 ENST00000536635.6 ENST00000085219.10 ENST00000544992.6 ENST00000419549.6 |
CD22
|
CD22 molecule |
| chr5_+_321695 | 0.20 |
ENST00000684583.1
ENST00000514523.1 |
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr6_-_48111132 | 0.19 |
ENST00000398738.3
ENST00000679966.1 ENST00000339488.9 |
PTCHD4
|
patched domain containing 4 |
| chr21_+_7748899 | 0.19 |
ENST00000623661.1
|
SMIM11B
|
small integral membrane protein 11B |
| chr2_+_29097705 | 0.19 |
ENST00000401605.5
ENST00000401617.6 |
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr1_+_203682734 | 0.19 |
ENST00000341360.6
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr7_-_97872420 | 0.19 |
ENST00000444334.5
ENST00000422745.5 ENST00000451771.5 ENST00000175506.8 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr9_-_114682061 | 0.19 |
ENST00000612244.5
|
TEX48
|
testis expressed 48 |
| chr15_+_76336755 | 0.19 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr8_-_13514744 | 0.18 |
ENST00000316609.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr10_+_100462969 | 0.18 |
ENST00000343737.6
|
WNT8B
|
Wnt family member 8B |
| chr17_-_36176636 | 0.18 |
ENST00000611257.5
ENST00000616006.1 |
TBC1D3B
|
TBC1 domain family member 3B |
| chr2_-_162074394 | 0.18 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4 |
| chr13_+_113158642 | 0.17 |
ENST00000375547.7
ENST00000342783.5 |
PROZ
|
protein Z, vitamin K dependent plasma glycoprotein |
| chr12_-_9999176 | 0.17 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr7_+_80638662 | 0.17 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr7_-_72828171 | 0.17 |
ENST00000612372.4
ENST00000610600.1 ENST00000620995.5 |
TYW1B
|
tRNA-yW synthesizing protein 1 homolog B |
| chr18_-_23662810 | 0.16 |
ENST00000322980.13
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr3_+_139935176 | 0.16 |
ENST00000458420.7
|
CLSTN2
|
calsyntenin 2 |
| chr3_-_197299067 | 0.16 |
ENST00000663148.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr10_+_116591010 | 0.16 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr10_+_116590956 | 0.16 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr12_+_56521798 | 0.16 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr11_+_116829898 | 0.16 |
ENST00000227667.8
ENST00000375345.3 |
APOC3
|
apolipoprotein C3 |
| chr7_-_102356444 | 0.16 |
ENST00000563237.3
|
SPDYE6
|
speedy/RINGO cell cycle regulator family member E6 |
| chr4_+_186227501 | 0.16 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr7_+_80638633 | 0.16 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr6_-_134052594 | 0.15 |
ENST00000275230.6
|
SLC2A12
|
solute carrier family 2 member 12 |
| chr19_-_49867251 | 0.15 |
ENST00000631020.2
ENST00000596014.5 ENST00000636994.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr8_-_13276491 | 0.14 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr4_+_678189 | 0.14 |
ENST00000507804.1
|
MYL5
|
myosin light chain 5 |
| chr7_+_69824565 | 0.14 |
ENST00000643587.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr17_-_69150062 | 0.14 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr19_+_6364543 | 0.14 |
ENST00000646643.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr17_+_44142015 | 0.14 |
ENST00000245382.6
|
HROB
|
homologous recombination factor with OB-fold |
| chr17_+_44141899 | 0.14 |
ENST00000319977.8
ENST00000585683.6 |
HROB
|
homologous recombination factor with OB-fold |
| chr5_+_148312416 | 0.14 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr5_+_136058849 | 0.13 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr8_-_21913671 | 0.13 |
ENST00000523932.1
ENST00000276420.9 |
DOK2
|
docking protein 2 |
| chr11_+_57338344 | 0.13 |
ENST00000263314.3
ENST00000616487.4 |
P2RX3
|
purinergic receptor P2X 3 |
| chr16_+_84768246 | 0.13 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr10_-_49539112 | 0.13 |
ENST00000355832.10
ENST00000447839.7 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr6_+_167291309 | 0.13 |
ENST00000230256.8
|
UNC93A
|
unc-93 homolog A |
| chr17_-_41060109 | 0.13 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr2_-_85418421 | 0.13 |
ENST00000409275.1
|
CAPG
|
capping actin protein, gelsolin like |
| chr12_+_10307818 | 0.13 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr10_-_49539015 | 0.13 |
ENST00000681659.1
ENST00000680107.1 |
ERCC6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr11_+_14904997 | 0.13 |
ENST00000523376.5
|
CALCB
|
calcitonin related polypeptide beta |
| chr10_+_35605337 | 0.12 |
ENST00000321660.2
|
GJD4
|
gap junction protein delta 4 |
| chr17_+_59220446 | 0.12 |
ENST00000284116.9
ENST00000581140.5 ENST00000581276.5 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_-_197299281 | 0.12 |
ENST00000419354.5
ENST00000667104.1 ENST00000658701.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_104854104 | 0.12 |
ENST00000361360.4
|
POU3F3
|
POU class 3 homeobox 3 |
| chr2_+_219498857 | 0.12 |
ENST00000684669.1
ENST00000373917.7 ENST00000435316.6 ENST00000313597.10 ENST00000358215.8 ENST00000373908.5 ENST00000622191.2 ENST00000683752.1 ENST00000341142.8 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr12_+_112978562 | 0.12 |
ENST00000680122.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr1_+_207752046 | 0.12 |
ENST00000367042.6
ENST00000322875.8 ENST00000322918.9 ENST00000354848.5 ENST00000357714.5 ENST00000358170.6 ENST00000367041.5 ENST00000367047.5 ENST00000360212.6 ENST00000480003.5 |
CD46
|
CD46 molecule |
| chr6_+_167291329 | 0.12 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A |
| chr7_-_76541459 | 0.11 |
ENST00000632547.1
|
SPDYE16
|
speedy/RINGO cell cycle regulator family member E16 |
| chr1_+_62194831 | 0.11 |
ENST00000498273.2
|
L1TD1
|
LINE1 type transposase domain containing 1 |
| chr5_+_153491174 | 0.11 |
ENST00000521843.6
|
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chrX_-_124963768 | 0.11 |
ENST00000371130.7
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr17_-_40867200 | 0.11 |
ENST00000647902.1
ENST00000251643.5 |
KRT12
|
keratin 12 |
| chr16_+_77788554 | 0.11 |
ENST00000302536.3
|
VAT1L
|
vesicle amine transport 1 like |
| chr19_-_37210484 | 0.11 |
ENST00000527838.5
ENST00000591492.5 ENST00000532828.7 |
ZNF585B
|
zinc finger protein 585B |
| chr11_+_60455839 | 0.10 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chrX_+_48383516 | 0.10 |
ENST00000620320.4
ENST00000595689.3 |
SSX4
|
SSX family member 4 |
| chr3_-_36739791 | 0.10 |
ENST00000416516.2
|
DCLK3
|
doublecortin like kinase 3 |
| chr18_+_63752935 | 0.10 |
ENST00000425392.5
ENST00000336429.6 |
SERPINB7
|
serpin family B member 7 |
| chr6_-_63319943 | 0.10 |
ENST00000622415.1
ENST00000370658.9 ENST00000485906.6 ENST00000370657.9 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr5_-_157166262 | 0.10 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71 member B |
| chr1_-_207911384 | 0.10 |
ENST00000356522.4
|
CD34
|
CD34 molecule |
| chr2_+_108588286 | 0.10 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr1_-_207911027 | 0.10 |
ENST00000310833.12
|
CD34
|
CD34 molecule |
| chr12_-_55958582 | 0.10 |
ENST00000549404.5
|
PMEL
|
premelanosome protein |
| chr14_+_61529005 | 0.10 |
ENST00000556347.1
|
ENSG00000258989.1
|
novel protein |
| chr8_-_94475044 | 0.10 |
ENST00000297592.5
ENST00000336148.10 |
RAD54B
|
RAD54 homolog B |
| chr5_+_40909490 | 0.10 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr18_+_63777773 | 0.09 |
ENST00000447428.5
ENST00000546027.5 |
SERPINB7
|
serpin family B member 7 |
| chr4_+_40749909 | 0.09 |
ENST00000316607.5
ENST00000381782.7 |
NSUN7
|
NOP2/Sun RNA methyltransferase family member 7 |
| chr16_-_71230676 | 0.09 |
ENST00000321489.9
ENST00000539973.5 ENST00000288168.14 ENST00000545267.1 ENST00000541601.5 ENST00000393567.7 ENST00000538248.5 |
HYDIN
|
HYDIN axonemal central pair apparatus protein |
| chr22_+_22343185 | 0.09 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr13_+_112968496 | 0.09 |
ENST00000397030.5
|
MCF2L
|
MCF.2 cell line derived transforming sequence like |
| chr3_-_197183849 | 0.09 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr3_-_119660580 | 0.09 |
ENST00000493094.6
ENST00000264231.7 ENST00000468801.1 |
POPDC2
|
popeye domain containing 2 |
| chr5_-_115602416 | 0.09 |
ENST00000427199.3
|
TICAM2
|
toll like receptor adaptor molecule 2 |
| chr4_+_158315309 | 0.09 |
ENST00000460056.6
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr4_-_145180496 | 0.09 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr13_+_95433593 | 0.09 |
ENST00000376873.7
|
CLDN10
|
claudin 10 |
| chr6_-_10412367 | 0.09 |
ENST00000379608.9
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr7_+_151341764 | 0.09 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
| chr8_-_13514821 | 0.09 |
ENST00000276297.9
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr1_-_155991198 | 0.09 |
ENST00000673475.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr4_+_70197924 | 0.08 |
ENST00000514097.5
|
ODAM
|
odontogenic, ameloblast associated |
| chr9_-_5185628 | 0.08 |
ENST00000381641.4
ENST00000649639.1 |
INSL6
|
insulin like 6 |
| chr20_-_46684467 | 0.08 |
ENST00000372121.5
|
SLC13A3
|
solute carrier family 13 member 3 |
| chr5_-_142698004 | 0.08 |
ENST00000407758.5
ENST00000441680.6 ENST00000419524.6 ENST00000621536.4 |
FGF1
|
fibroblast growth factor 1 |
| chr16_-_47459130 | 0.08 |
ENST00000565940.6
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr5_-_135895834 | 0.08 |
ENST00000274520.2
|
IL9
|
interleukin 9 |
| chr12_+_122078740 | 0.08 |
ENST00000319080.12
|
MLXIP
|
MLX interacting protein |
| chr6_-_137173644 | 0.08 |
ENST00000296980.7
ENST00000349184.8 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor subunit alpha 2 |
| chr1_+_180928133 | 0.08 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr19_+_49388243 | 0.08 |
ENST00000447857.8
|
KASH5
|
KASH domain containing 5 |
| chr5_-_59586393 | 0.08 |
ENST00000505453.1
ENST00000360047.9 |
PDE4D
|
phosphodiesterase 4D |
| chr1_-_201399302 | 0.07 |
ENST00000633953.1
ENST00000391967.7 |
LAD1
|
ladinin 1 |
| chrX_-_131289266 | 0.07 |
ENST00000370910.5
ENST00000370901.4 ENST00000361420.8 ENST00000370903.8 |
IGSF1
|
immunoglobulin superfamily member 1 |
| chr16_+_72054477 | 0.07 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr13_+_53028806 | 0.07 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr11_-_18791563 | 0.07 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5 |
| chr2_+_178320228 | 0.07 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr14_-_106154113 | 0.07 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr3_-_125120813 | 0.07 |
ENST00000430155.6
|
SLC12A8
|
solute carrier family 12 member 8 |
| chr22_-_30246739 | 0.07 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr6_-_130970428 | 0.07 |
ENST00000529208.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr21_-_14658812 | 0.07 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr14_+_22314435 | 0.07 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40 |
| chr17_+_60422483 | 0.07 |
ENST00000269127.5
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr3_-_197183806 | 0.07 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr8_+_127735597 | 0.07 |
ENST00000651626.1
|
MYC
|
MYC proto-oncogene, bHLH transcription factor |
| chr4_+_113049616 | 0.07 |
ENST00000504454.5
ENST00000357077.9 ENST00000394537.7 ENST00000672779.1 ENST00000264366.10 |
ANK2
|
ankyrin 2 |
| chr6_+_25652201 | 0.06 |
ENST00000612225.4
ENST00000377961.3 |
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr12_+_6840800 | 0.06 |
ENST00000541978.5
ENST00000229264.8 ENST00000435982.6 |
GNB3
|
G protein subunit beta 3 |
| chr2_-_160200289 | 0.06 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chrX_+_100666854 | 0.06 |
ENST00000640282.1
|
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr11_-_18791768 | 0.06 |
ENST00000358540.7
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5 |
| chr4_+_113049479 | 0.06 |
ENST00000671727.1
ENST00000671762.1 ENST00000672366.1 ENST00000672502.1 ENST00000672045.1 ENST00000672251.1 ENST00000672854.1 |
ANK2
|
ankyrin 2 |
| chr16_-_87317364 | 0.06 |
ENST00000567970.2
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr9_+_84668485 | 0.06 |
ENST00000359847.4
ENST00000395882.6 ENST00000376208.6 ENST00000376213.6 |
NTRK2
|
neurotrophic receptor tyrosine kinase 2 |
| chr22_-_35824373 | 0.06 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr1_+_84144260 | 0.06 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_171066936 | 0.06 |
ENST00000453628.1
ENST00000434911.6 |
TLK1
|
tousled like kinase 1 |
| chr2_+_191245473 | 0.06 |
ENST00000304164.8
|
MYO1B
|
myosin IB |
| chr8_+_39934955 | 0.06 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr17_-_9791586 | 0.06 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr5_-_43040311 | 0.06 |
ENST00000616064.2
|
ANXA2R
|
annexin A2 receptor |
| chr1_+_160739265 | 0.06 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr12_-_44875647 | 0.06 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2 |
| chr17_-_3298360 | 0.06 |
ENST00000323404.2
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr8_+_120445402 | 0.06 |
ENST00000305949.6
|
MTBP
|
MDM2 binding protein |
| chr19_+_4198075 | 0.06 |
ENST00000262970.9
|
ANKRD24
|
ankyrin repeat domain 24 |
| chrX_-_53434341 | 0.06 |
ENST00000375298.4
ENST00000375304.9 ENST00000684692.1 ENST00000168216.11 |
HSD17B10
|
hydroxysteroid 17-beta dehydrogenase 10 |
| chr3_+_98353854 | 0.06 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4 |
| chr2_-_85398734 | 0.06 |
ENST00000453973.5
|
CAPG
|
capping actin protein, gelsolin like |
| chr1_-_201399525 | 0.06 |
ENST00000367313.4
|
LAD1
|
ladinin 1 |
| chr14_+_67720842 | 0.06 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr2_+_11534039 | 0.06 |
ENST00000381486.7
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr17_+_7336502 | 0.05 |
ENST00000158762.8
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr2_-_160200251 | 0.05 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr12_-_69699455 | 0.05 |
ENST00000266661.8
|
BEST3
|
bestrophin 3 |
| chr8_+_58411333 | 0.05 |
ENST00000399598.7
|
UBXN2B
|
UBX domain protein 2B |
| chr10_+_5692792 | 0.05 |
ENST00000645567.1
|
TASOR2
|
transcription activation suppressor family member 2 |
| chr8_+_41490396 | 0.05 |
ENST00000518270.5
ENST00000520817.5 |
GOLGA7
|
golgin A7 |
| chr14_+_63204859 | 0.05 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr12_-_44876294 | 0.05 |
ENST00000429094.7
ENST00000551601.5 ENST00000549027.5 ENST00000452445.6 |
NELL2
|
neural EGFL like 2 |
| chr19_-_53824288 | 0.05 |
ENST00000324134.11
ENST00000391773.6 ENST00000391775.7 ENST00000345770.9 ENST00000391772.1 |
NLRP12
|
NLR family pyrin domain containing 12 |
| chr11_+_844406 | 0.05 |
ENST00000397404.5
|
TSPAN4
|
tetraspanin 4 |
| chr6_-_131000722 | 0.05 |
ENST00000528282.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr10_-_88851809 | 0.05 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr5_-_170297746 | 0.04 |
ENST00000046794.10
|
LCP2
|
lymphocyte cytosolic protein 2 |
| chr18_+_21242224 | 0.04 |
ENST00000424526.6
|
GREB1L
|
GREB1 like retinoic acid receptor coactivator |
| chr22_-_29061831 | 0.04 |
ENST00000216071.5
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr1_+_2073986 | 0.04 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta |
| chr2_-_40453438 | 0.04 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr17_-_40885232 | 0.04 |
ENST00000167588.4
|
KRT20
|
keratin 20 |
| chr5_+_120464236 | 0.04 |
ENST00000407149.7
ENST00000379551.2 |
PRR16
|
proline rich 16 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.2 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:0060621 | regulation of high-density lipoprotein particle clearance(GO:0010982) negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.2 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0072564 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.1 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.3 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.0 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0071438 | intercellular canaliculus(GO:0046581) invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |