Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PAX8
Z-value: 0.52
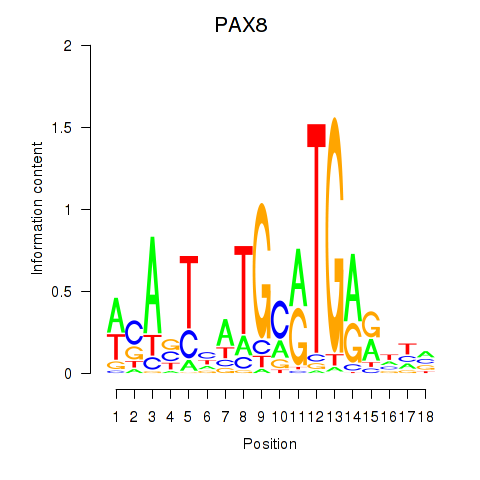
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.18 | PAX8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg38_v1_chr2_-_113241683_113241697 | 0.08 | 6.8e-01 | Click! |
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_37920602 | 0.70 |
ENST00000199448.9
ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr19_-_55160668 | 0.63 |
ENST00000588076.1
|
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr10_+_84173793 | 0.62 |
ENST00000372126.4
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr6_+_52420107 | 0.57 |
ENST00000636489.1
ENST00000637089.1 ENST00000637353.1 ENST00000637263.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr12_-_55842950 | 0.52 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19 |
| chr19_-_51034840 | 0.52 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr6_-_24935942 | 0.52 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr19_-_35528221 | 0.50 |
ENST00000588674.5
ENST00000452271.7 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr19_-_51034727 | 0.47 |
ENST00000525263.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr1_-_24415035 | 0.47 |
ENST00000374409.5
|
STPG1
|
sperm tail PG-rich repeat containing 1 |
| chr14_+_67720842 | 0.44 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr17_+_40287861 | 0.43 |
ENST00000209728.9
ENST00000580824.5 ENST00000577249.1 ENST00000649662.1 |
CDC6
|
cell division cycle 6 |
| chr19_-_51034892 | 0.42 |
ENST00000319590.8
ENST00000250351.4 |
KLK12
|
kallikrein related peptidase 12 |
| chr19_+_49665958 | 0.42 |
ENST00000594157.5
ENST00000600947.5 ENST00000598306.2 |
BCL2L12
|
BCL2 like 12 |
| chr17_+_63998344 | 0.42 |
ENST00000577953.5
ENST00000582540.5 ENST00000579184.5 ENST00000425164.7 ENST00000412177.6 ENST00000583891.5 ENST00000580752.1 |
PRR29
|
proline rich 29 |
| chr10_-_27983103 | 0.42 |
ENST00000672841.1
|
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr3_-_128123765 | 0.41 |
ENST00000322623.10
|
RUVBL1
|
RuvB like AAA ATPase 1 |
| chr7_-_97872394 | 0.41 |
ENST00000455086.5
ENST00000394308.8 ENST00000453600.5 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr6_+_31158518 | 0.39 |
ENST00000376255.4
ENST00000376257.8 |
TCF19
|
transcription factor 19 |
| chr9_-_136996555 | 0.39 |
ENST00000494426.2
|
CLIC3
|
chloride intracellular channel 3 |
| chr2_+_119544420 | 0.39 |
ENST00000413369.8
|
CFAP221
|
cilia and flagella associated protein 221 |
| chr12_-_56741535 | 0.38 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr19_-_51034993 | 0.36 |
ENST00000684732.1
|
KLK12
|
kallikrein related peptidase 12 |
| chr11_-_105139750 | 0.36 |
ENST00000530950.2
|
CARD18
|
caspase recruitment domain family member 18 |
| chr2_-_233854566 | 0.36 |
ENST00000432087.5
ENST00000441687.5 ENST00000414924.5 |
HJURP
|
Holliday junction recognition protein |
| chr6_+_29306626 | 0.33 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr8_+_39934955 | 0.32 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr20_-_3781440 | 0.31 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr2_-_79086847 | 0.31 |
ENST00000454188.5
|
REG1B
|
regenerating family member 1 beta |
| chr8_-_100105619 | 0.31 |
ENST00000523287.5
ENST00000519092.5 |
RGS22
|
regulator of G protein signaling 22 |
| chr21_-_34511243 | 0.30 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr18_+_63775369 | 0.30 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr14_-_106622837 | 0.30 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr21_-_42395943 | 0.30 |
ENST00000398405.5
|
TMPRSS3
|
transmembrane serine protease 3 |
| chr12_-_6851245 | 0.29 |
ENST00000540683.1
ENST00000229265.10 ENST00000535406.5 ENST00000422785.7 ENST00000538862.7 |
CDCA3
|
cell division cycle associated 3 |
| chr19_+_49665926 | 0.29 |
ENST00000246784.8
|
BCL2L12
|
BCL2 like 12 |
| chr12_-_54258275 | 0.29 |
ENST00000552562.1
|
CBX5
|
chromobox 5 |
| chr5_-_88877967 | 0.28 |
ENST00000508610.5
ENST00000636294.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr19_+_35140022 | 0.28 |
ENST00000588081.5
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr21_-_7789509 | 0.28 |
ENST00000646133.1
|
SMIM34B
|
small integral membrane protein 34B |
| chr1_+_182450132 | 0.28 |
ENST00000294854.13
|
RGSL1
|
regulator of G protein signaling like 1 |
| chr19_-_7702124 | 0.27 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr1_+_110451132 | 0.27 |
ENST00000271331.4
|
PROK1
|
prokineticin 1 |
| chr18_-_60372767 | 0.26 |
ENST00000299766.5
|
MC4R
|
melanocortin 4 receptor |
| chr1_+_171257930 | 0.26 |
ENST00000354841.4
|
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr11_-_26572254 | 0.26 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr17_+_58169401 | 0.25 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr4_+_36281591 | 0.25 |
ENST00000639862.2
ENST00000357504.7 |
DTHD1
|
death domain containing 1 |
| chr12_-_49900250 | 0.25 |
ENST00000552669.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr12_-_52680398 | 0.24 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr1_+_155033824 | 0.24 |
ENST00000295542.6
ENST00000423025.6 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr7_+_90245207 | 0.23 |
ENST00000497910.5
|
CFAP69
|
cilia and flagella associated protein 69 |
| chr19_+_41796616 | 0.22 |
ENST00000344550.4
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr7_+_20615653 | 0.22 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr10_+_12349685 | 0.21 |
ENST00000378845.5
|
CAMK1D
|
calcium/calmodulin dependent protein kinase ID |
| chr10_+_24449426 | 0.21 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr6_+_24126186 | 0.21 |
ENST00000378478.5
ENST00000378491.9 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr19_-_7702139 | 0.21 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr22_-_50085414 | 0.20 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr15_-_55498317 | 0.20 |
ENST00000568310.1
ENST00000565113.5 ENST00000448430.6 ENST00000457155.6 |
DNAAF4-CCPG1
DNAAF4
|
DNAAF4-CCPG1 readthrough (NMD candidate) dynein axonemal assembly factor 4 |
| chr19_+_9087061 | 0.20 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr9_-_21240002 | 0.20 |
ENST00000380222.4
|
IFNA14
|
interferon alpha 14 |
| chr12_+_124993633 | 0.20 |
ENST00000341446.9
ENST00000671775.2 |
BRI3BP
|
BRI3 binding protein |
| chr9_-_21228222 | 0.20 |
ENST00000413767.2
|
IFNA17
|
interferon alpha 17 |
| chr5_+_122129597 | 0.19 |
ENST00000514925.1
|
ENSG00000250803.6
|
novel zinc finger protein |
| chrX_+_140091415 | 0.19 |
ENST00000453380.2
|
ENSG00000230707.2
|
novel protein (LOC389895) |
| chr22_-_50085331 | 0.18 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr12_+_26195647 | 0.18 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr11_+_113314569 | 0.18 |
ENST00000429951.5
ENST00000442859.5 ENST00000531164.5 ENST00000529850.5 ENST00000529221.6 ENST00000525965.5 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr9_-_114074969 | 0.18 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr12_-_86256267 | 0.18 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chrX_-_101052054 | 0.17 |
ENST00000372939.5
ENST00000372935.5 |
TRMT2B
|
tRNA methyltransferase 2 homolog B |
| chr2_+_218659656 | 0.17 |
ENST00000443791.5
ENST00000359273.8 ENST00000392111.7 ENST00000392109.5 ENST00000392110.6 ENST00000423377.5 ENST00000412366.5 |
BCS1L
|
BCS1 homolog, ubiquinol-cytochrome c reductase complex chaperone |
| chr17_+_58166982 | 0.17 |
ENST00000545221.2
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr7_-_99679987 | 0.17 |
ENST00000222982.8
ENST00000439761.3 ENST00000339843.6 |
CYP3A5
|
cytochrome P450 family 3 subfamily A member 5 |
| chr9_+_136979042 | 0.16 |
ENST00000446677.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr2_+_218659749 | 0.16 |
ENST00000439945.5
ENST00000431802.5 |
BCS1L
|
BCS1 homolog, ubiquinol-cytochrome c reductase complex chaperone |
| chr6_-_29375291 | 0.16 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3 |
| chr18_+_45825666 | 0.16 |
ENST00000389474.8
|
SIGLEC15
|
sialic acid binding Ig like lectin 15 |
| chr1_+_100352451 | 0.15 |
ENST00000361544.11
ENST00000370124.8 ENST00000336454.5 |
CDC14A
|
cell division cycle 14A |
| chr13_+_76952467 | 0.15 |
ENST00000449753.2
|
ACOD1
|
aconitate decarboxylase 1 |
| chr16_-_30762052 | 0.15 |
ENST00000543610.6
ENST00000545825.1 ENST00000541260.5 |
CCDC189
|
coiled-coil domain containing 189 |
| chr19_-_38812936 | 0.15 |
ENST00000307751.9
ENST00000594209.1 |
LGALS4
|
galectin 4 |
| chr1_-_99766620 | 0.15 |
ENST00000646001.2
|
FRRS1
|
ferric chelate reductase 1 |
| chr2_-_135530561 | 0.14 |
ENST00000536680.5
ENST00000401392.5 |
ZRANB3
|
zinc finger RANBP2-type containing 3 |
| chr14_+_21978440 | 0.14 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr6_+_72212887 | 0.14 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_14803649 | 0.14 |
ENST00000330828.3
|
C12orf60
|
chromosome 12 open reading frame 60 |
| chr6_+_72212802 | 0.14 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr6_+_116877236 | 0.13 |
ENST00000332958.3
|
RFX6
|
regulatory factor X6 |
| chr20_+_59996335 | 0.13 |
ENST00000244049.7
ENST00000350849.10 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr11_+_7605719 | 0.13 |
ENST00000530181.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr12_+_75480800 | 0.13 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr8_+_22057857 | 0.13 |
ENST00000517305.4
ENST00000265800.9 ENST00000517418.5 |
DMTN
|
dematin actin binding protein |
| chr2_+_87748087 | 0.13 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr9_+_135075422 | 0.13 |
ENST00000371799.8
ENST00000277415.15 |
OLFM1
|
olfactomedin 1 |
| chr17_-_76027212 | 0.13 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr1_-_10976215 | 0.12 |
ENST00000520253.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr6_-_32816910 | 0.12 |
ENST00000447394.1
ENST00000438763.7 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr12_-_57016517 | 0.12 |
ENST00000441881.5
ENST00000458521.7 |
TAC3
|
tachykinin precursor 3 |
| chr1_+_159439722 | 0.12 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr6_-_48111132 | 0.12 |
ENST00000398738.3
ENST00000679966.1 ENST00000339488.9 |
PTCHD4
|
patched domain containing 4 |
| chr12_-_9208388 | 0.12 |
ENST00000261336.7
|
PZP
|
PZP alpha-2-macroglobulin like |
| chr1_-_152580511 | 0.12 |
ENST00000368787.4
|
LCE3D
|
late cornified envelope 3D |
| chr18_-_31684504 | 0.12 |
ENST00000383131.3
ENST00000237019.11 ENST00000306851.10 |
B4GALT6
|
beta-1,4-galactosyltransferase 6 |
| chr12_-_55842927 | 0.11 |
ENST00000322569.9
ENST00000409200.7 |
MMP19
|
matrix metallopeptidase 19 |
| chr3_-_114759115 | 0.11 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_8769602 | 0.11 |
ENST00000316793.8
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr19_-_55280063 | 0.11 |
ENST00000593263.5
ENST00000433386.7 |
HSPBP1
|
HSPA (Hsp70) binding protein 1 |
| chr11_+_60971777 | 0.11 |
ENST00000542157.5
ENST00000433107.6 ENST00000352009.9 ENST00000452451.6 |
CD6
|
CD6 molecule |
| chr14_+_75985747 | 0.11 |
ENST00000679083.1
ENST00000314067.11 ENST00000238628.10 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 |
| chr11_+_61229583 | 0.11 |
ENST00000544899.1
|
PGA4
|
pepsinogen A4 |
| chr1_-_58546693 | 0.11 |
ENST00000456980.5
ENST00000482274.2 ENST00000453710.1 ENST00000371226.8 ENST00000419242.5 ENST00000426139.5 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr15_+_89088417 | 0.11 |
ENST00000569550.5
ENST00000565066.5 ENST00000565973.5 ENST00000352732.10 |
ABHD2
|
abhydrolase domain containing 2, acylglycerol lipase |
| chr22_-_37007798 | 0.11 |
ENST00000402860.7
|
TEX33
|
testis expressed 33 |
| chr22_-_37007818 | 0.10 |
ENST00000405091.6
|
TEX33
|
testis expressed 33 |
| chr22_-_37007844 | 0.10 |
ENST00000381821.2
|
TEX33
|
testis expressed 33 |
| chr10_+_133527355 | 0.10 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr3_+_190388120 | 0.10 |
ENST00000456423.2
ENST00000264734.3 |
CLDN16
|
claudin 16 |
| chr16_-_3717505 | 0.10 |
ENST00000538171.5
ENST00000246957.10 |
TRAP1
|
TNF receptor associated protein 1 |
| chr1_+_160739239 | 0.10 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr19_+_17226662 | 0.10 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr12_+_71664352 | 0.10 |
ENST00000547843.1
|
THAP2
|
THAP domain containing 2 |
| chr19_-_55280194 | 0.10 |
ENST00000588971.1
ENST00000255631.9 ENST00000587551.1 |
HSPBP1
|
HSPA (Hsp70) binding protein 1 |
| chr4_-_144140725 | 0.10 |
ENST00000360771.8
|
GYPA
|
glycophorin A (MNS blood group) |
| chr17_+_69414690 | 0.09 |
ENST00000590474.7
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_-_57680260 | 0.09 |
ENST00000434992.1
|
NLRP2B
|
NLR family pyrin domain containing 2B |
| chr3_-_149377637 | 0.09 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chrX_-_72239022 | 0.09 |
ENST00000373657.2
ENST00000334463.4 |
ERCC6L
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr1_+_89524819 | 0.09 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr11_+_9461003 | 0.09 |
ENST00000438144.6
ENST00000396602.7 ENST00000526657.5 ENST00000299606.6 ENST00000534265.5 ENST00000412390.6 |
ZNF143
|
zinc finger protein 143 |
| chr16_-_28506826 | 0.09 |
ENST00000356897.1
|
IL27
|
interleukin 27 |
| chr2_+_90209873 | 0.09 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr7_-_76626127 | 0.09 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_-_153541765 | 0.09 |
ENST00000368718.5
|
S100A5
|
S100 calcium binding protein A5 |
| chr3_-_38794042 | 0.09 |
ENST00000643924.1
|
SCN10A
|
sodium voltage-gated channel alpha subunit 10 |
| chr2_+_27148997 | 0.09 |
ENST00000296096.6
|
TCF23
|
transcription factor 23 |
| chr3_+_157436842 | 0.08 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr5_+_140647802 | 0.08 |
ENST00000417647.7
ENST00000507593.5 ENST00000508301.5 |
IK
|
IK cytokine |
| chr5_-_113434978 | 0.08 |
ENST00000390666.4
|
TSSK1B
|
testis specific serine kinase 1B |
| chr12_-_95996302 | 0.08 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr15_-_43493076 | 0.08 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr18_+_63542365 | 0.08 |
ENST00000269491.6
ENST00000382768.2 |
SERPINB12
|
serpin family B member 12 |
| chr17_-_63931354 | 0.08 |
ENST00000647774.1
|
ENSG00000285947.1
|
novel protein |
| chr4_-_127965930 | 0.08 |
ENST00000641447.1
ENST00000296468.8 ENST00000641134.1 ENST00000641147.1 ENST00000641178.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr4_+_118888918 | 0.08 |
ENST00000434046.6
|
SYNPO2
|
synaptopodin 2 |
| chr2_-_26641363 | 0.08 |
ENST00000288861.5
|
CIB4
|
calcium and integrin binding family member 4 |
| chr6_+_36885848 | 0.08 |
ENST00000355190.7
ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr20_-_43726989 | 0.08 |
ENST00000373003.2
|
GTSF1L
|
gametocyte specific factor 1 like |
| chr13_+_75788838 | 0.07 |
ENST00000497947.6
|
LMO7
|
LIM domain 7 |
| chr12_+_75480745 | 0.07 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr1_+_89524871 | 0.07 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr17_-_62806632 | 0.07 |
ENST00000583803.1
ENST00000456609.6 |
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr2_-_24328113 | 0.07 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr14_+_63761836 | 0.07 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chrX_-_33211540 | 0.07 |
ENST00000357033.9
|
DMD
|
dystrophin |
| chr7_-_144835981 | 0.07 |
ENST00000360057.7
ENST00000378099.7 ENST00000639328.1 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr5_+_90640718 | 0.06 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr3_-_138132381 | 0.06 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr7_+_140696696 | 0.06 |
ENST00000247866.9
ENST00000464566.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr13_+_72782225 | 0.06 |
ENST00000615625.1
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr17_+_7014774 | 0.06 |
ENST00000439424.6
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr6_-_138107412 | 0.06 |
ENST00000421351.4
|
PERP
|
p53 apoptosis effector related to PMP22 |
| chr19_+_51225059 | 0.06 |
ENST00000436584.6
ENST00000421133.6 ENST00000262262.5 ENST00000391796.7 |
CD33
|
CD33 molecule |
| chr3_-_16605416 | 0.06 |
ENST00000399444.7
|
DAZL
|
deleted in azoospermia like |
| chr6_+_160121809 | 0.06 |
ENST00000366963.9
|
SLC22A1
|
solute carrier family 22 member 1 |
| chr10_-_43397220 | 0.06 |
ENST00000477108.5
ENST00000544000.5 |
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_+_71664281 | 0.05 |
ENST00000308086.3
|
THAP2
|
THAP domain containing 2 |
| chr22_-_42070778 | 0.05 |
ENST00000396398.8
ENST00000403363.5 ENST00000402937.1 |
NAGA
|
alpha-N-acetylgalactosaminidase |
| chr1_-_182604379 | 0.05 |
ENST00000367558.6
|
RGS16
|
regulator of G protein signaling 16 |
| chr1_-_169734064 | 0.05 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr13_+_27270814 | 0.05 |
ENST00000241463.5
|
RASL11A
|
RAS like family 11 member A |
| chr18_-_23662810 | 0.05 |
ENST00000322980.13
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr12_+_77830886 | 0.05 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr5_+_149141573 | 0.05 |
ENST00000506113.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr7_+_140696665 | 0.05 |
ENST00000476279.5
ENST00000461457.1 ENST00000465506.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr7_+_140697144 | 0.05 |
ENST00000476470.5
ENST00000471136.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr3_+_44712634 | 0.05 |
ENST00000449836.5
ENST00000296091.8 ENST00000436624.7 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr13_+_49110309 | 0.05 |
ENST00000398316.7
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr4_+_118888829 | 0.05 |
ENST00000448416.6
ENST00000307142.9 ENST00000429713.7 |
SYNPO2
|
synaptopodin 2 |
| chr11_+_123358416 | 0.05 |
ENST00000638157.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr17_+_32444379 | 0.05 |
ENST00000578213.5
ENST00000649012.1 ENST00000457654.6 ENST00000579451.1 ENST00000261712.8 |
PSMD11
|
proteasome 26S subunit, non-ATPase 11 |
| chr4_+_124491 | 0.05 |
ENST00000510175.6
ENST00000609714.1 |
ZNF718
|
zinc finger protein 718 |
| chr3_-_65622073 | 0.05 |
ENST00000621418.4
ENST00000611645.4 |
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_3595831 | 0.05 |
ENST00000399759.7
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1 |
| chr4_-_79408198 | 0.05 |
ENST00000358842.5
|
GK2
|
glycerol kinase 2 |
| chr7_+_140696956 | 0.04 |
ENST00000460088.5
ENST00000472695.5 |
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr3_-_161372821 | 0.04 |
ENST00000617024.1
ENST00000359175.8 |
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr19_-_52786742 | 0.04 |
ENST00000338230.3
ENST00000648973.1 |
ZNF600
|
zinc finger protein 600 |
| chr7_+_7566866 | 0.04 |
ENST00000405785.5
ENST00000340080.9 ENST00000433635.1 |
MIOS
|
meiosis regulator for oocyte development |
| chr15_+_67128103 | 0.04 |
ENST00000558894.5
|
SMAD3
|
SMAD family member 3 |
| chr10_+_74176537 | 0.04 |
ENST00000672394.1
|
ADK
|
adenosine kinase |
| chr17_-_75271205 | 0.04 |
ENST00000649805.1
|
MIF4GD
|
MIF4G domain containing |
| chr3_-_190122317 | 0.04 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr19_-_3500625 | 0.04 |
ENST00000672935.1
|
DOHH
|
deoxyhypusine hydroxylase |
| chr11_+_60455839 | 0.04 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr10_+_120851341 | 0.04 |
ENST00000263461.11
|
WDR11
|
WD repeat domain 11 |
| chr8_-_7056729 | 0.04 |
ENST00000330590.4
|
DEFA5
|
defensin alpha 5 |
| chr4_+_8580387 | 0.04 |
ENST00000382487.5
|
GPR78
|
G protein-coupled receptor 78 |
| chrX_+_130401962 | 0.04 |
ENST00000305536.11
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein X-linked 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.5 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 0.4 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.2 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 0.5 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.6 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.3 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.7 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.0 | 0.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.2 | GO:0072573 | propionate metabolic process(GO:0019541) cellular response to progesterone stimulus(GO:0071393) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 1.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.4 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 2.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.3 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |