Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
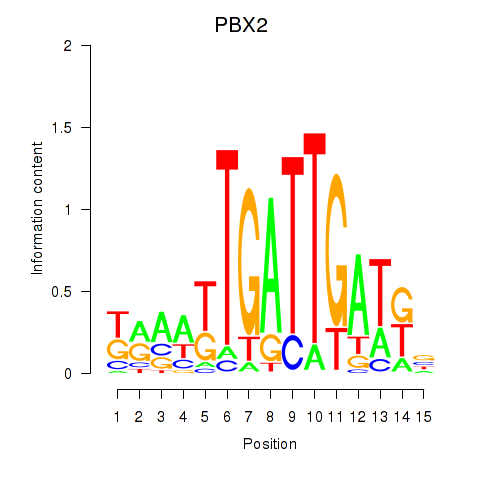
Results for PBX2
Z-value: 0.25
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.12 | PBX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg38_v1_chr6_-_32190170_32190214 | 0.60 | 4.7e-04 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_31217511 | 0.92 |
ENST00000403897.4
|
CAPN14
|
calpain 14 |
| chr14_-_55191534 | 0.66 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr12_-_27971970 | 0.60 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr12_-_27972725 | 0.48 |
ENST00000545234.6
|
PTHLH
|
parathyroid hormone like hormone |
| chr3_+_190615308 | 0.45 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr15_+_67125707 | 0.35 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr2_+_233692881 | 0.27 |
ENST00000305139.11
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr14_-_23155302 | 0.24 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr3_+_189171948 | 0.22 |
ENST00000345063.8
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr11_+_62665305 | 0.18 |
ENST00000532971.2
|
CSKMT
|
citrate synthase lysine methyltransferase |
| chr11_-_55936400 | 0.16 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chrX_+_46837034 | 0.16 |
ENST00000218340.4
|
RP2
|
RP2 activator of ARL3 GTPase |
| chr22_-_30246739 | 0.15 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr4_+_37960397 | 0.15 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr2_-_99301195 | 0.14 |
ENST00000308528.9
|
LYG1
|
lysozyme g1 |
| chr7_+_134843884 | 0.14 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr1_+_206865620 | 0.14 |
ENST00000367098.6
|
IL20
|
interleukin 20 |
| chr10_-_44978789 | 0.14 |
ENST00000448778.1
ENST00000298295.4 |
DEPP1
|
DEPP1 autophagy regulator |
| chr2_+_112275588 | 0.14 |
ENST00000409871.6
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr1_+_61952283 | 0.14 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr12_+_4590075 | 0.14 |
ENST00000540757.6
|
DYRK4
|
dual specificity tyrosine phosphorylation regulated kinase 4 |
| chr9_+_107306459 | 0.13 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr11_-_111923722 | 0.13 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chrX_+_120604199 | 0.12 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr14_+_64388296 | 0.11 |
ENST00000554739.5
ENST00000554768.6 ENST00000652179.1 ENST00000652337.1 ENST00000557370.3 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 |
| chr11_-_104898670 | 0.11 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene) |
| chr17_-_30824665 | 0.11 |
ENST00000324238.7
|
CRLF3
|
cytokine receptor like factor 3 |
| chr2_+_203867943 | 0.10 |
ENST00000295854.10
ENST00000487393.1 ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr10_+_122560751 | 0.09 |
ENST00000338354.10
ENST00000664692.1 ENST00000653442.1 ENST00000664974.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr17_-_1229706 | 0.09 |
ENST00000574139.7
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr1_+_101237009 | 0.08 |
ENST00000305352.7
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr10_+_122560679 | 0.08 |
ENST00000657942.1
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_+_203867764 | 0.08 |
ENST00000648405.2
|
CTLA4
|
cytotoxic T-lymphocyte associated protein 4 |
| chr2_+_233693659 | 0.07 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr10_+_122560639 | 0.07 |
ENST00000344338.7
ENST00000330163.8 ENST00000652446.2 ENST00000666315.1 ENST00000368955.7 ENST00000368909.7 ENST00000368956.6 ENST00000619379.1 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_+_20447065 | 0.07 |
ENST00000272233.6
|
RHOB
|
ras homolog family member B |
| chr3_-_52830664 | 0.07 |
ENST00000266041.9
ENST00000406595.5 ENST00000485816.5 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr2_+_33436304 | 0.07 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr10_-_73808616 | 0.06 |
ENST00000299641.8
|
NDST2
|
N-deacetylase and N-sulfotransferase 2 |
| chr3_-_129440007 | 0.06 |
ENST00000503197.5
ENST00000249910.5 ENST00000507208.1 ENST00000393278.6 |
MBD4
|
methyl-CpG binding domain 4, DNA glycosylase |
| chr5_+_55024250 | 0.06 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chrX_+_120604084 | 0.06 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr18_-_75208417 | 0.06 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr7_-_151520080 | 0.06 |
ENST00000496004.5
|
RHEB
|
Ras homolog, mTORC1 binding |
| chr2_-_264024 | 0.06 |
ENST00000403712.6
ENST00000356150.10 ENST00000626873.2 ENST00000405430.5 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr3_+_160225409 | 0.05 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr1_+_61952036 | 0.05 |
ENST00000646453.1
ENST00000635137.1 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr7_-_112939773 | 0.04 |
ENST00000297145.9
|
BMT2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr3_-_129439925 | 0.04 |
ENST00000429544.7
|
MBD4
|
methyl-CpG binding domain 4, DNA glycosylase |
| chr1_-_114780624 | 0.04 |
ENST00000060969.6
ENST00000369528.9 |
SIKE1
|
suppressor of IKBKE 1 |
| chr9_+_122510802 | 0.04 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor family 1 subfamily J member 2 |
| chr2_+_44168866 | 0.03 |
ENST00000282412.9
ENST00000409432.7 ENST00000378551.6 ENST00000345249.8 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent 1B |
| chrX_-_31178220 | 0.03 |
ENST00000681026.1
|
DMD
|
dystrophin |
| chr18_-_75209126 | 0.03 |
ENST00000322342.4
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr7_-_33100886 | 0.02 |
ENST00000448915.1
|
RP9
|
RP9 pre-mRNA splicing factor |
| chr4_+_99574812 | 0.02 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chrX_-_31178149 | 0.02 |
ENST00000679437.1
|
DMD
|
dystrophin |
| chr14_+_22516273 | 0.00 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr5_+_141392616 | 0.00 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 1.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 0.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004487 | formate-tetrahydrofolate ligase activity(GO:0004329) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |