Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for PITX2
Z-value: 0.37
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
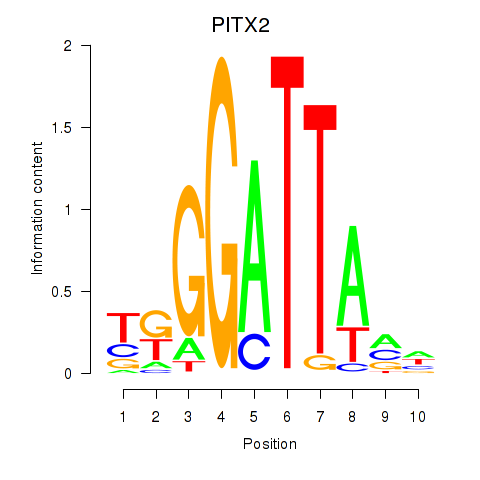
PITX2
|
ENSG00000164093.17 | PITX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg38_v1_chr4_-_110636963_110637042, hg38_v1_chr4_-_110641920_110642123, hg38_v1_chr4_-_110623051_110623085 | -0.44 | 1.5e-02 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5227063 | 1.73 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr9_+_34458752 | 1.68 |
ENST00000614641.4
ENST00000242317.9 ENST00000437363.5 |
DNAI1
|
dynein axonemal intermediate chain 1 |
| chr13_-_35855627 | 1.68 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr22_-_30471986 | 0.94 |
ENST00000401751.5
ENST00000402286.5 ENST00000403066.5 ENST00000215812.9 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr13_-_35855758 | 0.70 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr6_-_111483700 | 0.64 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr10_-_60141004 | 0.57 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr12_-_89656051 | 0.57 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr12_-_89656093 | 0.55 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr1_-_165445088 | 0.46 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr14_+_75522427 | 0.44 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr1_-_165445220 | 0.43 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr22_-_17258235 | 0.41 |
ENST00000649310.1
ENST00000649746.1 |
ADA2
|
adenosine deaminase 2 |
| chr5_+_38258373 | 0.36 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr11_-_119346695 | 0.34 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chr7_-_128775793 | 0.34 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive |
| chr12_-_54981838 | 0.33 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr14_+_75522531 | 0.31 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr11_-_133532493 | 0.31 |
ENST00000524381.6
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr1_+_202010575 | 0.29 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr16_-_11976611 | 0.28 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chr3_-_191282383 | 0.27 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr12_+_69739370 | 0.27 |
ENST00000550536.5
ENST00000362025.9 |
RAB3IP
|
RAB3A interacting protein |
| chr10_-_48274567 | 0.26 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr17_+_4740005 | 0.26 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr13_+_73058993 | 0.25 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chr17_+_4740042 | 0.25 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr6_-_41705813 | 0.22 |
ENST00000419574.6
ENST00000445214.2 |
TFEB
|
transcription factor EB |
| chr11_+_61755372 | 0.20 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chrM_+_9207 | 0.20 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr11_-_96343170 | 0.20 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr1_+_21977014 | 0.15 |
ENST00000337107.11
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr17_-_4739866 | 0.15 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr8_+_48008409 | 0.15 |
ENST00000523432.5
ENST00000521346.5 ENST00000523111.7 ENST00000517630.5 |
UBE2V2
|
ubiquitin conjugating enzyme E2 V2 |
| chr4_-_67754335 | 0.15 |
ENST00000420975.2
ENST00000226413.5 |
GNRHR
|
gonadotropin releasing hormone receptor |
| chr13_-_94479671 | 0.14 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr19_+_2476118 | 0.14 |
ENST00000215631.9
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA damage inducible beta |
| chr15_+_40929338 | 0.12 |
ENST00000249749.7
|
DLL4
|
delta like canonical Notch ligand 4 |
| chr11_-_119346655 | 0.12 |
ENST00000360167.4
|
MFRP
|
membrane frizzled-related protein |
| chr3_+_154121366 | 0.11 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr7_+_74453790 | 0.11 |
ENST00000265755.7
ENST00000424337.7 ENST00000455841.6 |
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr6_-_53148822 | 0.11 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr1_-_30757767 | 0.11 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr5_-_150758497 | 0.10 |
ENST00000521533.1
ENST00000424236.5 |
DCTN4
|
dynactin subunit 4 |
| chr16_-_67980483 | 0.10 |
ENST00000268793.6
ENST00000672962.1 |
DPEP3
|
dipeptidase 3 |
| chr16_+_28711417 | 0.09 |
ENST00000395587.5
ENST00000569690.5 ENST00000331666.11 ENST00000564243.5 ENST00000566866.5 |
EIF3C
|
eukaryotic translation initiation factor 3 subunit C |
| chr7_+_30284574 | 0.06 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2 |
| chr21_-_33542841 | 0.06 |
ENST00000381831.7
ENST00000381839.7 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr10_+_17951906 | 0.04 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chr9_-_14300231 | 0.03 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr10_+_17951885 | 0.02 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr10_+_17951825 | 0.02 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr19_-_41436439 | 0.01 |
ENST00000594660.5
|
DMAC2
|
distal membrane arm assembly complex 2 |
| chrX_+_15749848 | 0.00 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.6 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 1.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 2.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.8 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.9 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |