Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
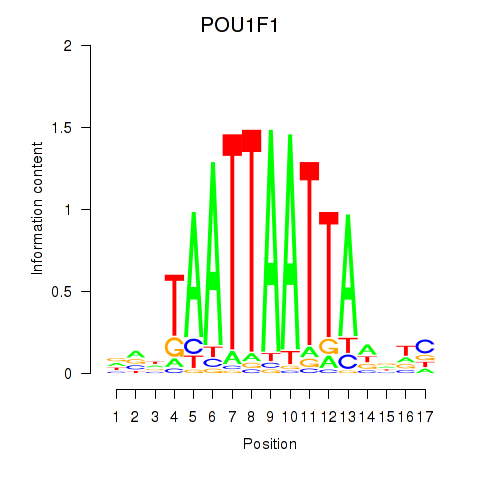
Results for POU1F1
Z-value: 0.49
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.12 | POU1F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg38_v1_chr3_-_87276462_87276494 | -0.36 | 5.3e-02 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33283205 | 2.37 |
ENST00000253354.2
|
BPIFB1
|
BPI fold containing family B member 1 |
| chr10_-_13001705 | 1.84 |
ENST00000378825.5
|
CCDC3
|
coiled-coil domain containing 3 |
| chr15_-_56465130 | 1.61 |
ENST00000260453.4
|
MNS1
|
meiosis specific nuclear structural 1 |
| chr4_-_16083695 | 1.60 |
ENST00000510224.5
|
PROM1
|
prominin 1 |
| chr4_-_16083714 | 1.59 |
ENST00000508167.5
|
PROM1
|
prominin 1 |
| chr19_+_55836532 | 1.18 |
ENST00000301295.11
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr2_+_227871618 | 1.08 |
ENST00000309931.3
ENST00000440997.1 |
DAW1
|
dynein assembly factor with WD repeats 1 |
| chr11_+_102047422 | 1.04 |
ENST00000434758.7
ENST00000526781.5 ENST00000534360.1 |
CFAP300
|
cilia and flagella associated protein 300 |
| chr3_-_167380270 | 0.92 |
ENST00000392764.5
ENST00000675490.1 ENST00000474464.5 ENST00000392766.6 ENST00000485651.5 |
ZBBX
|
zinc finger B-box domain containing |
| chr13_-_85799400 | 0.90 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr16_-_75556214 | 0.89 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr16_-_28623560 | 0.83 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr14_-_106470788 | 0.83 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr4_-_99435336 | 0.75 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_130953898 | 0.75 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr22_+_23145366 | 0.71 |
ENST00000341989.9
ENST00000263116.8 |
RAB36
|
RAB36, member RAS oncogene family |
| chr4_-_99435134 | 0.67 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_69653223 | 0.66 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr3_-_191282383 | 0.64 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr11_-_26572254 | 0.63 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr2_-_27489716 | 0.61 |
ENST00000260570.8
ENST00000675690.1 |
IFT172
|
intraflagellar transport 172 |
| chr4_-_99435396 | 0.61 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_+_14583076 | 0.60 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr4_-_69860138 | 0.59 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr9_+_96928310 | 0.56 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr19_+_48695952 | 0.51 |
ENST00000522966.2
ENST00000425340.3 ENST00000391876.5 |
FUT2
|
fucosyltransferase 2 |
| chr1_-_60073750 | 0.49 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr12_-_10130241 | 0.48 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr11_-_26572130 | 0.45 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr16_-_28623330 | 0.44 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr12_-_10130143 | 0.44 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr6_-_49744434 | 0.39 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chrX_-_11290478 | 0.39 |
ENST00000380717.7
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr3_+_138621207 | 0.38 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_-_53759634 | 0.38 |
ENST00000557913.5
ENST00000360509.10 |
WDR72
|
WD repeat domain 72 |
| chr16_-_2329687 | 0.37 |
ENST00000567910.1
|
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr1_+_11273188 | 0.36 |
ENST00000376810.6
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr12_-_10130082 | 0.36 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr6_-_49744378 | 0.34 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr11_-_63608542 | 0.34 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr18_+_58341038 | 0.33 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_183590876 | 0.32 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr13_-_48533165 | 0.31 |
ENST00000430805.6
ENST00000544492.5 ENST00000544904.3 |
RCBTB2
|
RCC1 and BTB domain containing protein 2 |
| chr10_+_92834594 | 0.31 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_25052732 | 0.31 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr17_+_20073538 | 0.29 |
ENST00000681116.1
ENST00000680572.1 ENST00000680604.1 ENST00000681875.1 ENST00000679058.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr8_+_75539862 | 0.29 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr6_+_26365215 | 0.29 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr8_+_24294044 | 0.29 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr19_+_4198075 | 0.28 |
ENST00000262970.9
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr7_+_117014881 | 0.28 |
ENST00000422922.5
ENST00000432298.5 |
ST7
|
suppression of tumorigenicity 7 |
| chr13_+_36819214 | 0.28 |
ENST00000255476.3
|
RFXAP
|
regulatory factor X associated protein |
| chr6_+_26365176 | 0.27 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr3_-_65622073 | 0.27 |
ENST00000621418.4
ENST00000611645.4 |
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_-_28282086 | 0.27 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chr8_+_75539893 | 0.27 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr12_-_52367478 | 0.27 |
ENST00000257901.7
|
KRT85
|
keratin 85 |
| chr4_-_117085541 | 0.26 |
ENST00000310754.5
|
TRAM1L1
|
translocation associated membrane protein 1 like 1 |
| chr7_+_92057602 | 0.26 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr5_+_148312416 | 0.25 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr12_+_69359673 | 0.25 |
ENST00000548020.5
ENST00000549685.5 ENST00000247843.7 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr17_+_68259164 | 0.25 |
ENST00000448504.6
|
ARSG
|
arylsulfatase G |
| chr4_+_75556048 | 0.25 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr4_+_112647059 | 0.23 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator |
| chr1_-_53945661 | 0.23 |
ENST00000194214.10
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr18_+_6834473 | 0.23 |
ENST00000581099.5
ENST00000419673.6 ENST00000531294.5 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr10_-_27240505 | 0.23 |
ENST00000375888.5
ENST00000676732.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr18_-_55423757 | 0.23 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chrX_+_109535775 | 0.22 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr1_-_53945584 | 0.22 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr6_+_29301701 | 0.22 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr1_+_40396766 | 0.22 |
ENST00000539317.2
|
SMAP2
|
small ArfGAP2 |
| chr1_-_53945567 | 0.22 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chrX_-_32155462 | 0.22 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr17_+_7015035 | 0.21 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr16_-_66550142 | 0.21 |
ENST00000417693.8
ENST00000299697.12 ENST00000451102.7 |
TK2
|
thymidine kinase 2 |
| chr3_+_16174628 | 0.20 |
ENST00000339732.10
|
GALNT15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr9_-_28670285 | 0.20 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr11_-_59810750 | 0.20 |
ENST00000300151.5
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr10_+_70404129 | 0.20 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr13_-_70108441 | 0.20 |
ENST00000377844.9
ENST00000545028.2 |
KLHL1
|
kelch like family member 1 |
| chr1_-_111488795 | 0.20 |
ENST00000472933.2
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chrX_+_56563569 | 0.19 |
ENST00000338222.7
|
UBQLN2
|
ubiquilin 2 |
| chr10_+_87357720 | 0.19 |
ENST00000412718.3
ENST00000381697.7 |
NUTM2D
|
NUT family member 2D |
| chr14_-_25010604 | 0.19 |
ENST00000550887.5
|
STXBP6
|
syntaxin binding protein 6 |
| chr2_+_233917371 | 0.19 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr7_+_130293134 | 0.19 |
ENST00000445470.6
ENST00000492072.5 ENST00000222482.10 ENST00000473956.5 ENST00000493259.5 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr17_-_41009124 | 0.19 |
ENST00000391588.3
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr6_+_26103922 | 0.19 |
ENST00000377803.4
|
H4C3
|
H4 clustered histone 3 |
| chr2_-_218166951 | 0.18 |
ENST00000295683.3
|
CXCR1
|
C-X-C motif chemokine receptor 1 |
| chr11_-_129024157 | 0.18 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr9_-_96778053 | 0.17 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr1_+_212791828 | 0.17 |
ENST00000532324.5
ENST00000530441.5 ENST00000526641.5 ENST00000531963.5 ENST00000366973.8 ENST00000366974.9 ENST00000526997.5 ENST00000488246.6 |
TATDN3
|
TatD DNase domain containing 3 |
| chr14_+_21918161 | 0.17 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr10_-_112183698 | 0.17 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr9_-_5833014 | 0.17 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr20_+_6006039 | 0.17 |
ENST00000452938.5
ENST00000378863.9 |
CRLS1
|
cardiolipin synthase 1 |
| chr3_-_161105224 | 0.16 |
ENST00000651254.1
ENST00000651178.1 ENST00000476999.6 ENST00000652596.1 ENST00000651305.1 ENST00000652111.1 ENST00000651292.1 ENST00000651282.1 ENST00000651380.1 ENST00000494173.7 ENST00000484127.5 ENST00000650733.1 ENST00000494818.6 ENST00000492353.5 ENST00000652143.1 ENST00000473142.5 ENST00000651147.1 ENST00000468268.5 ENST00000460353.2 ENST00000651953.1 ENST00000651972.1 ENST00000652730.1 ENST00000651460.1 ENST00000652059.1 ENST00000651509.1 ENST00000651801.1 ENST00000651686.1 ENST00000320474.10 ENST00000392781.7 ENST00000392779.6 ENST00000651791.1 ENST00000651117.1 ENST00000652032.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr12_+_38316753 | 0.16 |
ENST00000551464.1
ENST00000308742.9 |
ALG10B
|
ALG10 alpha-1,2-glucosyltransferase B |
| chr19_-_22010930 | 0.16 |
ENST00000601773.5
ENST00000397126.9 ENST00000609966.5 ENST00000601993.1 ENST00000599916.5 |
ZNF208
|
zinc finger protein 208 |
| chr9_-_111330224 | 0.16 |
ENST00000302681.3
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2 |
| chr9_-_34729482 | 0.16 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A |
| chr8_+_106726115 | 0.16 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr9_+_93234923 | 0.16 |
ENST00000411624.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr8_-_85341659 | 0.16 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr1_+_149782671 | 0.16 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chrX_+_151397163 | 0.15 |
ENST00000330374.7
|
VMA21
|
vacuolar ATPase assembly factor VMA21 |
| chr19_+_49513353 | 0.15 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr19_-_14835252 | 0.15 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr3_-_161105399 | 0.15 |
ENST00000652593.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr14_+_19743571 | 0.15 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr1_-_211134061 | 0.15 |
ENST00000639602.1
ENST00000638498.1 ENST00000367007.5 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr4_+_186069144 | 0.15 |
ENST00000513189.1
ENST00000296795.8 |
TLR3
|
toll like receptor 3 |
| chr10_+_125896549 | 0.15 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr1_-_92486916 | 0.15 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr1_-_248277976 | 0.14 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr12_+_34022462 | 0.14 |
ENST00000538927.1
ENST00000266483.7 |
ALG10
|
ALG10 alpha-1,2-glucosyltransferase |
| chr17_+_7014774 | 0.14 |
ENST00000439424.6
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr9_-_110337808 | 0.14 |
ENST00000374510.8
ENST00000374507.4 ENST00000423740.7 ENST00000374511.7 |
TXNDC8
|
thioredoxin domain containing 8 |
| chr16_-_66550112 | 0.14 |
ENST00000544898.6
ENST00000620035.5 ENST00000545043.6 |
TK2
|
thymidine kinase 2 |
| chr2_+_102473219 | 0.14 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr20_-_31390580 | 0.14 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr1_+_152970646 | 0.14 |
ENST00000328051.3
|
SPRR4
|
small proline rich protein 4 |
| chr15_+_48191648 | 0.14 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr6_+_122996227 | 0.14 |
ENST00000275162.10
|
CLVS2
|
clavesin 2 |
| chr9_+_108862255 | 0.13 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
| chr17_+_7252502 | 0.13 |
ENST00000570322.5
ENST00000576496.5 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr4_+_37453914 | 0.13 |
ENST00000381980.9
ENST00000508175.5 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr15_-_75455767 | 0.13 |
ENST00000360439.8
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_208025494 | 0.13 |
ENST00000457206.1
ENST00000427836.8 |
PLEKHM3
|
pleckstrin homology domain containing M3 |
| chr17_+_58169401 | 0.13 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr15_+_58410543 | 0.13 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr19_+_49513154 | 0.12 |
ENST00000426395.7
ENST00000600273.5 ENST00000599988.5 |
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr3_+_138621225 | 0.12 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_89954518 | 0.12 |
ENST00000556005.1
ENST00000555872.5 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr4_+_95091462 | 0.12 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr10_-_77637902 | 0.12 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr11_-_63144221 | 0.12 |
ENST00000417740.5
ENST00000612278.4 ENST00000326192.5 |
SLC22A24
|
solute carrier family 22 member 24 |
| chr17_+_46511511 | 0.12 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr15_-_64093746 | 0.12 |
ENST00000557835.5
ENST00000380290.7 ENST00000300030.8 ENST00000559950.1 |
CIAO2A
|
cytosolic iron-sulfur assembly component 2A |
| chr15_+_64387828 | 0.12 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr6_+_158312459 | 0.12 |
ENST00000367097.8
|
TULP4
|
TUB like protein 4 |
| chr11_-_18526885 | 0.12 |
ENST00000251968.4
ENST00000536719.5 |
TSG101
|
tumor susceptibility 101 |
| chr8_-_100105619 | 0.12 |
ENST00000523287.5
ENST00000519092.5 |
RGS22
|
regulator of G protein signaling 22 |
| chr9_-_113303271 | 0.12 |
ENST00000297894.5
ENST00000489339.2 |
RNF183
|
ring finger protein 183 |
| chr20_-_31390483 | 0.11 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr6_-_109381739 | 0.11 |
ENST00000504373.2
|
CD164
|
CD164 molecule |
| chr3_-_142000353 | 0.11 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr12_-_48570046 | 0.11 |
ENST00000301046.6
ENST00000549817.1 |
LALBA
|
lactalbumin alpha |
| chr19_-_10587219 | 0.11 |
ENST00000591240.5
ENST00000589684.5 ENST00000591676.1 ENST00000250244.11 ENST00000590923.5 |
AP1M2
|
adaptor related protein complex 1 subunit mu 2 |
| chr4_+_48831217 | 0.11 |
ENST00000510824.5
ENST00000425583.6 |
OCIAD1
|
OCIA domain containing 1 |
| chr15_+_65550819 | 0.11 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr17_+_7407838 | 0.11 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr11_+_73787853 | 0.11 |
ENST00000310614.12
ENST00000497094.6 ENST00000411840.6 ENST00000535277.5 ENST00000398483.7 ENST00000542303.5 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr17_-_4739866 | 0.11 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr7_+_138460238 | 0.11 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr2_+_102418642 | 0.11 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr6_+_12716554 | 0.11 |
ENST00000676159.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr15_-_66356672 | 0.10 |
ENST00000261881.9
|
TIPIN
|
TIMELESS interacting protein |
| chr14_+_88594406 | 0.10 |
ENST00000555900.5
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr10_-_50279715 | 0.10 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr18_-_77127935 | 0.10 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr16_+_81238682 | 0.10 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr2_-_151261839 | 0.10 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43 |
| chr19_-_34773184 | 0.10 |
ENST00000588760.1
ENST00000329285.13 ENST00000587354.6 |
ZNF599
|
zinc finger protein 599 |
| chr2_-_86105839 | 0.10 |
ENST00000263857.11
|
POLR1A
|
RNA polymerase I subunit A |
| chr10_-_30629741 | 0.10 |
ENST00000647634.2
ENST00000375318.4 |
LYZL2
|
lysozyme like 2 |
| chr16_-_66550091 | 0.10 |
ENST00000564917.5
ENST00000677420.1 |
TK2
|
thymidine kinase 2 |
| chrX_+_85003863 | 0.10 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr14_+_88594430 | 0.10 |
ENST00000406216.7
ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr9_+_101185029 | 0.10 |
ENST00000395056.2
|
PLPPR1
|
phospholipid phosphatase related 1 |
| chr14_+_88594395 | 0.10 |
ENST00000318308.10
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr16_-_66550005 | 0.10 |
ENST00000527284.6
|
TK2
|
thymidine kinase 2 |
| chr16_-_66549839 | 0.10 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chr3_-_157503339 | 0.10 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr19_+_9087061 | 0.09 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr3_-_157503375 | 0.09 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chrX_+_83861126 | 0.09 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr7_-_16833411 | 0.09 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr19_-_9219589 | 0.09 |
ENST00000641244.1
ENST00000641669.1 |
OR7D4
|
olfactory receptor family 7 subfamily D member 4 |
| chr5_+_129748091 | 0.09 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2 |
| chr5_-_93741587 | 0.09 |
ENST00000606183.4
|
POU5F2
|
POU domain class 5, transcription factor 2 |
| chr5_+_55853314 | 0.09 |
ENST00000354961.8
ENST00000297015.7 |
IL31RA
|
interleukin 31 receptor A |
| chr11_+_59511368 | 0.09 |
ENST00000641278.1
|
OR4D9
|
olfactory receptor family 4 subfamily D member 9 |
| chr6_+_12716801 | 0.09 |
ENST00000674595.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr5_+_43120883 | 0.08 |
ENST00000515326.5
ENST00000682664.1 |
ZNF131
|
zinc finger protein 131 |
| chr1_-_205121986 | 0.08 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr1_-_205121964 | 0.08 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr20_-_7940444 | 0.08 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr7_-_138981307 | 0.08 |
ENST00000440172.5
ENST00000422774.2 |
KIAA1549
|
KIAA1549 |
| chr14_-_80231052 | 0.08 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr5_-_56116946 | 0.08 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr4_-_152679984 | 0.08 |
ENST00000304385.8
ENST00000504064.1 |
TMEM154
|
transmembrane protein 154 |
| chr14_-_24609660 | 0.07 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chr2_-_219399981 | 0.07 |
ENST00000519905.1
ENST00000523282.5 ENST00000434339.5 ENST00000457935.5 |
DNPEP
|
aspartyl aminopeptidase |
| chr4_+_119027335 | 0.07 |
ENST00000627783.2
|
SYNPO2
|
synaptopodin 2 |
| chr14_-_89954659 | 0.07 |
ENST00000555070.1
ENST00000316738.12 ENST00000538485.6 ENST00000556609.5 |
ENSG00000259053.1
EFCAB11
|
novel transcript EF-hand calcium binding domain 11 |
| chr17_+_1771688 | 0.07 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr12_-_57826295 | 0.07 |
ENST00000549039.5
|
CTDSP2
|
CTD small phosphatase 2 |
| chr1_-_212791762 | 0.07 |
ENST00000626725.1
ENST00000366977.8 ENST00000366976.3 |
NSL1
|
NSL1 component of MIS12 kinetochore complex |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 3.2 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 0.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 2.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 2.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.4 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.2 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 1.7 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.9 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 1.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.7 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.0 | 0.1 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:2000397 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.5 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 3.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.6 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.3 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 3.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |