Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for POU2F2_POU3F1
Z-value: 0.90
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
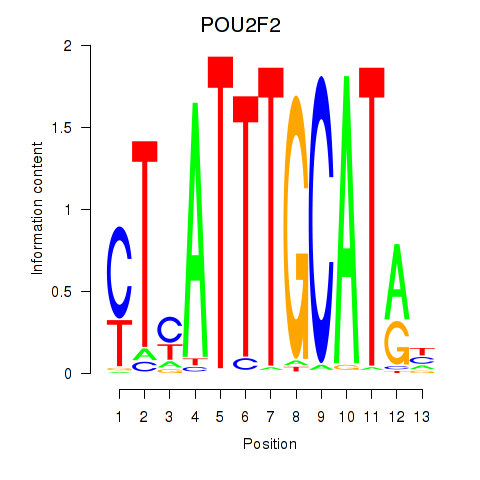
POU2F2
|
ENSG00000028277.21 | POU2F2 |
|
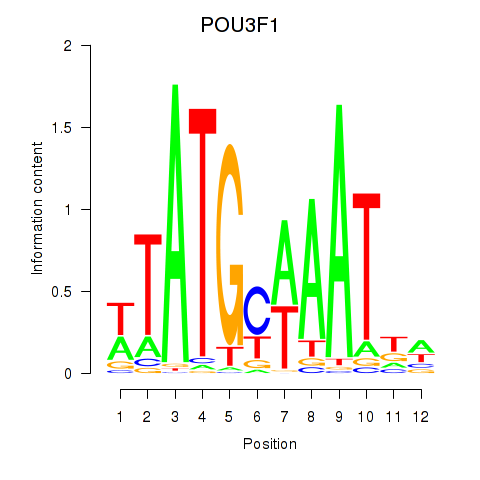
POU3F1
|
ENSG00000185668.8 | POU3F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg38_v1_chr19_-_42132391_42132455 | 0.34 | 6.6e-02 | Click! |
| POU3F1 | hg38_v1_chr1_-_38046785_38046802 | 0.12 | 5.2e-01 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26124161 | 2.42 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr3_+_63652663 | 2.41 |
ENST00000343837.8
ENST00000469440.5 |
SNTN
|
sentan, cilia apical structure protein |
| chr6_-_26123910 | 2.40 |
ENST00000314332.5
ENST00000396984.1 |
H2BC4
|
H2B clustered histone 4 |
| chr6_+_52420992 | 2.35 |
ENST00000636954.1
ENST00000636566.1 ENST00000638075.1 |
EFHC1
|
EF-hand domain containing 1 |
| chr6_-_27132750 | 2.30 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr1_-_149886652 | 2.23 |
ENST00000369155.3
|
H2BC21
|
H2B clustered histone 21 |
| chr1_-_183653307 | 2.20 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr6_-_26216673 | 2.07 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr9_-_72953047 | 2.02 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr14_-_106470788 | 1.89 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr1_-_228457855 | 1.86 |
ENST00000366695.3
|
H2AW
|
H2A.W histone |
| chr6_+_26216928 | 1.86 |
ENST00000303910.4
|
H2AC8
|
H2A clustered histone 8 |
| chr6_+_27133032 | 1.70 |
ENST00000359193.3
|
H2AC11
|
H2A clustered histone 11 |
| chrX_-_48919015 | 1.68 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr1_-_149842736 | 1.54 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr1_+_149851053 | 1.53 |
ENST00000607355.2
|
H2AC19
|
H2A clustered histone 19 |
| chr1_+_149886906 | 1.51 |
ENST00000331380.4
|
H2AC20
|
H2A clustered histone 20 |
| chr6_+_27147094 | 1.43 |
ENST00000377459.3
|
H2AC12
|
H2A clustered histone 12 |
| chr9_+_133459965 | 1.33 |
ENST00000540581.5
ENST00000542192.5 ENST00000291722.11 ENST00000316948.9 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr6_+_27865308 | 1.30 |
ENST00000613174.2
|
H2AC16
|
H2A clustered histone 16 |
| chr19_-_43619591 | 1.27 |
ENST00000598676.1
ENST00000300811.8 |
ZNF428
|
zinc finger protein 428 |
| chr15_-_19988117 | 1.23 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_-_238239958 | 1.16 |
ENST00000409182.1
ENST00000409002.7 ENST00000450098.1 ENST00000409356.1 ENST00000272937.10 ENST00000409160.7 ENST00000409574.1 |
HES6
|
hes family bHLH transcription factor 6 |
| chr11_-_5227063 | 1.15 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr12_+_48904116 | 1.15 |
ENST00000552942.5
ENST00000320516.5 |
CCDC65
|
coiled-coil domain containing 65 |
| chr6_+_26183750 | 1.14 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr6_-_27893175 | 1.08 |
ENST00000359611.4
|
H2AC17
|
H2A clustered histone 17 |
| chr9_-_81688354 | 1.05 |
ENST00000418319.5
|
TLE1
|
TLE family member 1, transcriptional corepressor |
| chr17_+_74737211 | 1.04 |
ENST00000392612.7
ENST00000392610.5 |
RAB37
|
RAB37, member RAS oncogene family |
| chr18_-_55422492 | 1.04 |
ENST00000561992.5
ENST00000630712.2 |
TCF4
|
transcription factor 4 |
| chr19_-_45584810 | 1.04 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr9_+_89605004 | 0.99 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr1_+_103655760 | 0.98 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A |
| chr1_-_149812359 | 0.98 |
ENST00000369167.2
ENST00000545683.1 |
H2BC18
|
H2B clustered histone 18 |
| chr17_-_75779758 | 0.97 |
ENST00000592643.5
ENST00000591890.5 ENST00000587171.1 ENST00000254810.8 ENST00000589599.5 |
H3-3B
|
H3.3 histone B |
| chr6_-_27146841 | 0.97 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr11_+_27040725 | 0.97 |
ENST00000529202.5
ENST00000263182.8 |
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr17_+_74737238 | 0.95 |
ENST00000392613.10
ENST00000613645.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr16_+_2969548 | 0.92 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr11_+_27041313 | 0.92 |
ENST00000528583.5
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr16_-_21303036 | 0.90 |
ENST00000219599.8
ENST00000576703.5 |
CRYM
|
crystallin mu |
| chr18_-_55422306 | 0.89 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr16_-_30096170 | 0.88 |
ENST00000566134.5
ENST00000565110.5 ENST00000398841.6 ENST00000398838.8 |
YPEL3
|
yippee like 3 |
| chr12_-_68302872 | 0.86 |
ENST00000539972.5
|
MDM1
|
Mdm1 nuclear protein |
| chr17_+_70104848 | 0.86 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr19_-_14884761 | 0.85 |
ENST00000642123.1
|
OR7A17
|
olfactory receptor family 7 subfamily A member 17 |
| chr15_+_83447328 | 0.82 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr12_-_7092422 | 0.81 |
ENST00000543835.5
ENST00000647956.2 ENST00000535233.6 |
C1R
|
complement C1r |
| chr14_+_64540734 | 0.80 |
ENST00000247207.7
|
HSPA2
|
heat shock protein family A (Hsp70) member 2 |
| chr15_+_83447411 | 0.79 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr8_+_55879818 | 0.78 |
ENST00000520220.6
ENST00000519728.6 |
LYN
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr1_+_59310071 | 0.78 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr6_+_27893411 | 0.78 |
ENST00000616182.2
|
H2BC17
|
H2B clustered histone 17 |
| chr16_+_2969307 | 0.78 |
ENST00000576565.1
ENST00000318782.9 |
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr16_+_2969270 | 0.77 |
ENST00000293978.12
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr6_-_27807916 | 0.77 |
ENST00000377401.3
|
H2BC13
|
H2B clustered histone 13 |
| chrX_-_24647300 | 0.77 |
ENST00000379144.7
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr19_-_17405554 | 0.74 |
ENST00000252593.7
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr1_+_61082398 | 0.74 |
ENST00000664149.1
|
NFIA
|
nuclear factor I A |
| chr17_-_19748355 | 0.74 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr7_-_130441136 | 0.73 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr19_+_17226597 | 0.72 |
ENST00000597836.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr16_+_33009175 | 0.71 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr1_+_202010575 | 0.71 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr11_+_27055215 | 0.71 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr1_-_154859841 | 0.70 |
ENST00000361147.8
|
KCNN3
|
potassium calcium-activated channel subfamily N member 3 |
| chr2_-_210171402 | 0.70 |
ENST00000281772.14
|
KANSL1L
|
KAT8 regulatory NSL complex subunit 1 like |
| chr19_+_17226662 | 0.69 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr3_-_112641128 | 0.69 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr13_-_35855758 | 0.69 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr19_+_17226218 | 0.69 |
ENST00000601529.5
ENST00000215061.9 ENST00000600232.5 |
OCEL1
|
occludin/ELL domain containing 1 |
| chrX_-_24647091 | 0.69 |
ENST00000356768.8
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_-_27814757 | 0.68 |
ENST00000333151.5
|
H2AC14
|
H2A clustered histone 14 |
| chr6_+_26158115 | 0.67 |
ENST00000377777.5
ENST00000289316.2 |
H2BC5
|
H2B clustered histone 5 |
| chr1_+_61081728 | 0.67 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr3_-_112846856 | 0.66 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr11_+_120236635 | 0.66 |
ENST00000260264.8
|
POU2F3
|
POU class 2 homeobox 3 |
| chr6_+_26272923 | 0.66 |
ENST00000377733.4
|
H2BC10
|
H2B clustered histone 10 |
| chr9_-_113303271 | 0.65 |
ENST00000297894.5
ENST00000489339.2 |
RNF183
|
ring finger protein 183 |
| chr2_-_210171327 | 0.65 |
ENST00000418791.5
ENST00000452086.5 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1 like |
| chr11_-_8594181 | 0.65 |
ENST00000358872.7
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr6_+_27815010 | 0.65 |
ENST00000621112.2
|
H2BC14
|
H2B clustered histone 14 |
| chr16_+_19417875 | 0.64 |
ENST00000542583.7
|
TMC5
|
transmembrane channel like 5 |
| chr4_-_109302707 | 0.63 |
ENST00000642955.1
|
COL25A1
|
collagen type XXV alpha 1 chain |
| chr1_+_167329044 | 0.61 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr6_-_81752671 | 0.59 |
ENST00000320172.11
ENST00000369754.7 ENST00000369756.3 |
TENT5A
|
terminal nucleotidyltransferase 5A |
| chr14_+_79279403 | 0.58 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr12_+_48904105 | 0.58 |
ENST00000266984.9
|
CCDC65
|
coiled-coil domain containing 65 |
| chr6_-_170291053 | 0.55 |
ENST00000366756.4
|
DLL1
|
delta like canonical Notch ligand 1 |
| chr17_-_19748285 | 0.54 |
ENST00000570414.1
ENST00000225740.11 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr2_+_96760857 | 0.54 |
ENST00000377075.3
|
CNNM4
|
cyclin and CBS domain divalent metal cation transport mediator 4 |
| chr17_-_19748341 | 0.54 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr16_-_33845229 | 0.53 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr18_+_46946821 | 0.51 |
ENST00000245121.9
ENST00000356157.12 |
KATNAL2
|
katanin catalytic subunit A1 like 2 |
| chr11_+_73308237 | 0.51 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr1_-_5992407 | 0.51 |
ENST00000378156.9
ENST00000622020.4 |
NPHP4
|
nephrocystin 4 |
| chr11_-_6030758 | 0.51 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr2_+_10721649 | 0.51 |
ENST00000381661.3
|
ATP6V1C2
|
ATPase H+ transporting V1 subunit C2 |
| chr11_+_537517 | 0.51 |
ENST00000270115.8
|
LRRC56
|
leucine rich repeat containing 56 |
| chr2_+_10721623 | 0.50 |
ENST00000272238.9
|
ATP6V1C2
|
ATPase H+ transporting V1 subunit C2 |
| chr4_-_185812209 | 0.50 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_14301069 | 0.50 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr14_-_106389858 | 0.49 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr7_+_98211431 | 0.49 |
ENST00000609256.2
|
BHLHA15
|
basic helix-loop-helix family member a15 |
| chr14_-_106154113 | 0.48 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr3_-_112641292 | 0.48 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr10_+_69451456 | 0.47 |
ENST00000373290.7
|
TSPAN15
|
tetraspanin 15 |
| chr16_-_11636357 | 0.47 |
ENST00000576334.1
ENST00000574848.5 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr10_+_104353820 | 0.47 |
ENST00000369704.8
|
CFAP58
|
cilia and flagella associated protein 58 |
| chr12_+_6944065 | 0.46 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr14_-_106165730 | 0.46 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr6_+_26251607 | 0.46 |
ENST00000619466.2
|
H2BC9
|
H2B clustered histone 9 |
| chr12_+_6943811 | 0.45 |
ENST00000544681.1
ENST00000537087.5 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr19_-_19033480 | 0.45 |
ENST00000452918.7
ENST00000600377.1 ENST00000337018.10 |
SUGP2
|
SURP and G-patch domain containing 2 |
| chr12_+_6944009 | 0.45 |
ENST00000229281.6
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr4_-_86101922 | 0.43 |
ENST00000472236.5
ENST00000641881.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_130440848 | 0.43 |
ENST00000675803.1
ENST00000223208.10 ENST00000343969.10 ENST00000471201.6 ENST00000675649.1 ENST00000675168.1 ENST00000469826.2 ENST00000334451.6 ENST00000675962.1 ENST00000675563.1 ENST00000480206.2 ENST00000489512.5 ENST00000676243.1 ENST00000674539.1 ENST00000675935.1 |
CEP41
|
centrosomal protein 41 |
| chr2_-_157325808 | 0.43 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr17_+_63773863 | 0.42 |
ENST00000578681.5
ENST00000583590.5 |
DDX42
|
DEAD-box helicase 42 |
| chr7_+_90709530 | 0.42 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr17_+_40015428 | 0.42 |
ENST00000394149.8
ENST00000225474.6 ENST00000331769.6 ENST00000394148.7 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 |
| chr12_-_55966018 | 0.41 |
ENST00000548803.5
ENST00000449260.6 ENST00000550447.5 ENST00000547137.5 ENST00000546543.5 ENST00000548747.6 ENST00000550464.5 |
PMEL
|
premelanosome protein |
| chr11_+_112175526 | 0.41 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr21_-_10649835 | 0.41 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr1_+_228458095 | 0.41 |
ENST00000620438.1
|
H2BU1
|
H2B.U histone 1 |
| chr17_+_20155989 | 0.41 |
ENST00000395530.6
ENST00000581399.6 ENST00000679819.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_-_23559490 | 0.40 |
ENST00000374561.6
|
ID3
|
inhibitor of DNA binding 3, HLH protein |
| chr6_-_26033609 | 0.40 |
ENST00000615868.2
|
H2AC4
|
H2A clustered histone 4 |
| chr16_+_85611401 | 0.39 |
ENST00000405402.6
|
GSE1
|
Gse1 coiled-coil protein |
| chr18_-_55585773 | 0.39 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr2_-_238288600 | 0.38 |
ENST00000254657.8
|
PER2
|
period circadian regulator 2 |
| chr3_-_18425295 | 0.38 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr6_+_138773747 | 0.38 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr11_-_62591500 | 0.37 |
ENST00000476907.6
ENST00000278279.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr11_-_62591554 | 0.37 |
ENST00000494385.1
ENST00000308436.11 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr17_+_63774144 | 0.37 |
ENST00000389924.7
ENST00000359353.9 |
DDX42
|
DEAD-box helicase 42 |
| chr6_+_10585748 | 0.37 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr7_-_113087720 | 0.37 |
ENST00000297146.7
|
GPR85
|
G protein-coupled receptor 85 |
| chr16_-_19886133 | 0.36 |
ENST00000568214.1
ENST00000569479.5 |
GPRC5B
|
G protein-coupled receptor class C group 5 member B |
| chr17_-_49764123 | 0.36 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr7_+_90709231 | 0.36 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr4_-_73223082 | 0.35 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr5_-_67196791 | 0.35 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr22_+_22431949 | 0.35 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr1_-_201115372 | 0.34 |
ENST00000458416.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chrX_+_70455093 | 0.34 |
ENST00000542398.1
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr6_+_27247690 | 0.34 |
ENST00000421826.6
ENST00000230582.8 |
PRSS16
|
serine protease 16 |
| chr5_+_102808057 | 0.34 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_86357722 | 0.34 |
ENST00000641341.1
ENST00000642038.1 ENST00000641116.1 ENST00000641767.1 ENST00000639242.1 ENST00000638313.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_-_26043704 | 0.34 |
ENST00000615966.2
|
H2BC3
|
H2B clustered histone 3 |
| chr18_-_55321640 | 0.34 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr4_-_25863537 | 0.33 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr10_-_102120246 | 0.33 |
ENST00000425280.2
|
LDB1
|
LIM domain binding 1 |
| chr11_+_112176364 | 0.33 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr12_-_31326171 | 0.33 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr17_+_40062810 | 0.33 |
ENST00000584985.5
ENST00000264637.8 |
THRA
|
thyroid hormone receptor alpha |
| chr14_-_106557465 | 0.33 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr5_+_73813518 | 0.32 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr1_-_156082412 | 0.32 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A |
| chr14_-_106803221 | 0.32 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr12_+_14774362 | 0.32 |
ENST00000544848.2
|
H2AJ
|
H2A.J histone |
| chr2_-_218002988 | 0.31 |
ENST00000682258.1
ENST00000446903.5 |
TNS1
|
tensin 1 |
| chr14_+_103107516 | 0.31 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3 like 4 |
| chr6_+_96562548 | 0.31 |
ENST00000541107.5
ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr6_-_142945028 | 0.31 |
ENST00000012134.7
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr7_-_120858066 | 0.31 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr6_-_55875583 | 0.30 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5 |
| chr15_-_40874216 | 0.30 |
ENST00000220507.5
|
RHOV
|
ras homolog family member V |
| chr6_-_142945160 | 0.30 |
ENST00000367603.8
|
HIVEP2
|
HIVEP zinc finger 2 |
| chrX_+_81202066 | 0.29 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr5_+_36608146 | 0.29 |
ENST00000381918.4
ENST00000513646.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr11_-_62601818 | 0.29 |
ENST00000278823.7
|
MTA2
|
metastasis associated 1 family member 2 |
| chr18_-_63644250 | 0.29 |
ENST00000341074.10
ENST00000436264.1 |
SERPINB4
|
serpin family B member 4 |
| chr19_+_40799155 | 0.29 |
ENST00000303961.9
|
EGLN2
|
egl-9 family hypoxia inducible factor 2 |
| chrX_-_64205680 | 0.28 |
ENST00000374869.8
|
AMER1
|
APC membrane recruitment protein 1 |
| chr15_-_64703199 | 0.28 |
ENST00000559753.1
ENST00000560258.6 ENST00000559912.2 ENST00000326005.10 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr16_-_73048104 | 0.28 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr6_-_149484965 | 0.28 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr5_-_19988179 | 0.27 |
ENST00000502796.5
ENST00000382275.6 ENST00000511273.1 |
CDH18
|
cadherin 18 |
| chr11_+_120240135 | 0.27 |
ENST00000543440.7
|
POU2F3
|
POU class 2 homeobox 3 |
| chr7_-_100586119 | 0.27 |
ENST00000310300.11
|
LRCH4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr6_+_143677935 | 0.27 |
ENST00000440869.6
ENST00000367582.7 ENST00000451827.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr10_-_127892930 | 0.26 |
ENST00000368671.4
|
CLRN3
|
clarin 3 |
| chrX_+_12975083 | 0.26 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr4_+_183905266 | 0.26 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr12_-_31326142 | 0.26 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr12_-_31326111 | 0.26 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr4_+_26320563 | 0.26 |
ENST00000361572.10
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_+_26224192 | 0.26 |
ENST00000634733.1
|
H3C6
|
H3 clustered histone 6 |
| chr7_-_120858303 | 0.25 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr5_+_76819022 | 0.25 |
ENST00000296677.5
|
F2RL1
|
F2R like trypsin receptor 1 |
| chr20_+_34704336 | 0.25 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr18_-_77127935 | 0.25 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr19_-_40425982 | 0.25 |
ENST00000357949.5
|
SERTAD1
|
SERTA domain containing 1 |
| chr17_-_65561137 | 0.25 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr6_-_35688935 | 0.24 |
ENST00000542713.1
|
FKBP5
|
FKBP prolyl isomerase 5 |
| chr17_+_65137408 | 0.24 |
ENST00000443584.7
ENST00000449996.7 |
RGS9
|
regulator of G protein signaling 9 |
| chr7_-_44082464 | 0.24 |
ENST00000335195.10
ENST00000395831.7 ENST00000414235.5 ENST00000242248.10 ENST00000452049.1 |
POLM
|
DNA polymerase mu |
| chr6_+_148342759 | 0.23 |
ENST00000367467.8
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr6_-_168079159 | 0.23 |
ENST00000283309.10
|
FRMD1
|
FERM domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.3 | 2.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 0.8 | GO:0070662 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) mast cell proliferation(GO:0070662) |
| 0.2 | 0.7 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.7 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.2 | 2.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.5 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.2 | 0.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.4 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.6 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.2 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.2 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.2 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.9 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.3 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 2.5 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 1.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 2.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.9 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.9 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0042323 | positive regulation of nucleobase-containing compound transport(GO:0032241) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.8 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 2.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 0.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 2.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 2.3 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 3.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 3.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 1.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 0.8 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 0.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.2 | 1.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 3.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 2.2 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 3.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |