Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
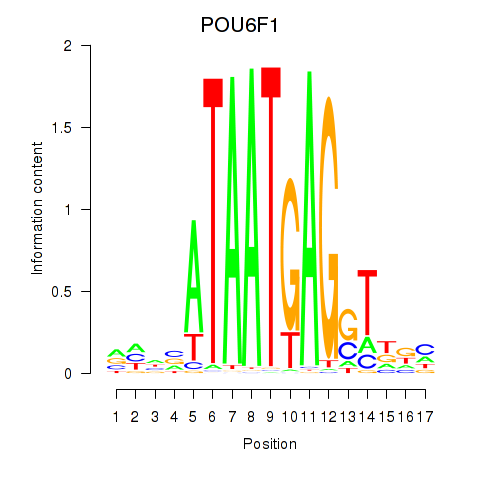
Results for POU6F1
Z-value: 0.33
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.17 | POU6F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F1 | hg38_v1_chr12_-_51217671_51217827 | 0.20 | 2.8e-01 | Click! |
Activity profile of POU6F1 motif
Sorted Z-values of POU6F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_151325665 | 0.82 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr6_+_27138588 | 0.56 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chr2_+_114461593 | 0.47 |
ENST00000409163.5
|
DPP10
|
dipeptidyl peptidase like 10 |
| chr5_+_90640718 | 0.41 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr12_-_30735014 | 0.40 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr1_-_153150884 | 0.35 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr10_+_11823348 | 0.32 |
ENST00000277570.10
ENST00000622831.4 |
PROSER2
|
proline and serine rich 2 |
| chr11_-_107858777 | 0.29 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr12_+_15546344 | 0.28 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr1_-_197146688 | 0.25 |
ENST00000294732.11
|
ASPM
|
assembly factor for spindle microtubules |
| chr4_+_105710809 | 0.24 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr6_+_26183750 | 0.24 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr1_-_197146620 | 0.24 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr14_+_94026314 | 0.20 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr13_+_97960192 | 0.20 |
ENST00000496368.6
ENST00000421861.7 ENST00000357602.7 |
IPO5
|
importin 5 |
| chr16_-_58684707 | 0.19 |
ENST00000564100.5
ENST00000570101.5 ENST00000562397.5 ENST00000219320.9 ENST00000564010.5 ENST00000570214.5 ENST00000563196.5 |
SLC38A7
|
solute carrier family 38 member 7 |
| chr2_+_202073282 | 0.18 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr6_-_132659178 | 0.18 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr14_+_22515623 | 0.18 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr2_+_190180930 | 0.17 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr2_-_216694794 | 0.17 |
ENST00000449583.1
|
IGFBP5
|
insulin like growth factor binding protein 5 |
| chr2_-_99301195 | 0.16 |
ENST00000308528.9
|
LYG1
|
lysozyme g1 |
| chr19_-_3786254 | 0.16 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr22_-_28306645 | 0.16 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr6_+_42155399 | 0.16 |
ENST00000623004.2
ENST00000372963.4 ENST00000654459.1 |
GUCA1ANB
GUCA1A
|
GUCA1A neighbor guanylate cyclase activator 1A |
| chr12_+_102120172 | 0.16 |
ENST00000327680.7
ENST00000541394.5 ENST00000543784.5 |
PARPBP
|
PARP1 binding protein |
| chr19_-_35563653 | 0.15 |
ENST00000262623.4
|
ATP4A
|
ATPase H+/K+ transporting subunit alpha |
| chr1_+_207325629 | 0.15 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr12_+_102120240 | 0.15 |
ENST00000537257.5
ENST00000392911.6 |
PARPBP
|
PARP1 binding protein |
| chr4_+_109827963 | 0.15 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chrX_+_43656289 | 0.14 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr20_+_59628609 | 0.14 |
ENST00000541461.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_-_138218491 | 0.14 |
ENST00000527246.3
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr19_-_3786408 | 0.14 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_-_3786363 | 0.13 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr2_-_165204042 | 0.13 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr11_+_2302119 | 0.13 |
ENST00000381121.7
|
TSPAN32
|
tetraspanin 32 |
| chr12_+_119668109 | 0.12 |
ENST00000229328.10
ENST00000630317.1 |
PRKAB1
|
protein kinase AMP-activated non-catalytic subunit beta 1 |
| chr13_+_30422487 | 0.12 |
ENST00000638137.1
ENST00000635918.2 |
UBE2L5
|
ubiquitin conjugating enzyme E2 L5 |
| chr21_-_31160904 | 0.12 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr18_+_63887698 | 0.12 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr22_-_21952827 | 0.11 |
ENST00000397495.8
ENST00000263212.10 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F |
| chr11_+_89924064 | 0.11 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr2_-_165203870 | 0.11 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr6_+_63521738 | 0.10 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr2_-_201697993 | 0.10 |
ENST00000428900.6
|
MPP4
|
membrane palmitoylated protein 4 |
| chr11_+_2301987 | 0.10 |
ENST00000612299.4
ENST00000182290.9 |
TSPAN32
|
tetraspanin 32 |
| chr6_+_25754699 | 0.09 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr9_+_128882119 | 0.09 |
ENST00000372600.9
ENST00000372599.7 |
LRRC8A
|
leucine rich repeat containing 8 VRAC subunit A |
| chr2_-_201698692 | 0.09 |
ENST00000315506.11
ENST00000359962.9 ENST00000620095.4 |
MPP4
|
membrane palmitoylated protein 4 |
| chr21_-_43060546 | 0.09 |
ENST00000430013.1
|
CBS
|
cystathionine beta-synthase |
| chr3_+_46354072 | 0.09 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr20_-_51802509 | 0.08 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr13_+_57631735 | 0.08 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr2_-_201698040 | 0.08 |
ENST00000396886.7
ENST00000409143.5 |
MPP4
|
membrane palmitoylated protein 4 |
| chr7_+_148590760 | 0.08 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr16_-_87317364 | 0.08 |
ENST00000567970.2
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr8_+_117135259 | 0.08 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr16_-_87317416 | 0.08 |
ENST00000253461.8
ENST00000618367.4 |
C16orf95
|
chromosome 16 open reading frame 95 |
| chr12_-_7444139 | 0.08 |
ENST00000416109.2
ENST00000313599.8 |
CD163L1
|
CD163 molecule like 1 |
| chr7_+_141790217 | 0.07 |
ENST00000247883.5
|
TAS2R5
|
taste 2 receptor member 5 |
| chr1_+_176463164 | 0.07 |
ENST00000367661.7
ENST00000367662.5 |
PAPPA2
|
pappalysin 2 |
| chr2_-_201698628 | 0.07 |
ENST00000602867.1
ENST00000409474.8 |
MPP4
|
membrane palmitoylated protein 4 |
| chr12_-_91153149 | 0.07 |
ENST00000550758.1
|
DCN
|
decorin |
| chr14_-_70416973 | 0.07 |
ENST00000555276.5
ENST00000617124.4 |
COX16
SYNJ2BP-COX16
|
cytochrome c oxidase assembly factor COX16 SYNJ2BP-COX16 readthrough |
| chr16_+_15650221 | 0.07 |
ENST00000396354.6
ENST00000570727.5 ENST00000674581.1 ENST00000674995.1 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr12_+_10307818 | 0.07 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr5_-_16916400 | 0.07 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr22_+_22704265 | 0.07 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chrX_+_120604084 | 0.07 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr3_+_77039836 | 0.06 |
ENST00000461745.5
|
ROBO2
|
roundabout guidance receptor 2 |
| chr1_+_224929648 | 0.06 |
ENST00000430092.5
ENST00000366850.7 ENST00000400952.7 ENST00000682510.1 ENST00000366849.5 |
DNAH14
|
dynein axonemal heavy chain 14 |
| chr6_-_47042260 | 0.06 |
ENST00000371243.2
|
ADGRF1
|
adhesion G protein-coupled receptor F1 |
| chr8_+_117134989 | 0.06 |
ENST00000456015.7
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr6_+_50818857 | 0.06 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr11_-_4393650 | 0.06 |
ENST00000254436.8
|
TRIM21
|
tripartite motif containing 21 |
| chr1_+_224952670 | 0.06 |
ENST00000366848.5
ENST00000439375.6 |
DNAH14
|
dynein axonemal heavy chain 14 |
| chr11_+_85628573 | 0.05 |
ENST00000393375.5
ENST00000358867.11 ENST00000534341.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr2_+_202073249 | 0.05 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr2_+_190180835 | 0.05 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr15_-_84716153 | 0.05 |
ENST00000455959.7
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr18_+_9474994 | 0.05 |
ENST00000019317.8
|
RALBP1
|
ralA binding protein 1 |
| chr12_-_95003666 | 0.04 |
ENST00000327772.7
ENST00000547157.1 ENST00000684171.1 ENST00000547986.5 |
NDUFA12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr6_-_26234978 | 0.04 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr6_-_110815408 | 0.04 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr9_-_128881922 | 0.04 |
ENST00000320665.10
ENST00000436267.7 ENST00000302586.8 ENST00000651925.1 |
KYAT1
ENSG00000286112.1
|
kynurenine aminotransferase 1 kynurenine aminotransferase 1 |
| chr6_-_110815152 | 0.04 |
ENST00000413605.6
|
CDK19
|
cyclin dependent kinase 19 |
| chr15_-_84716063 | 0.04 |
ENST00000558217.5
ENST00000558196.1 ENST00000558134.5 |
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr2_+_11539833 | 0.04 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr3_-_108529322 | 0.04 |
ENST00000273353.4
|
MYH15
|
myosin heavy chain 15 |
| chr11_+_118527463 | 0.03 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chrX_-_100874332 | 0.03 |
ENST00000372960.8
|
NOX1
|
NADPH oxidase 1 |
| chr15_-_84716129 | 0.03 |
ENST00000268220.12
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr9_-_114806031 | 0.03 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15 |
| chr15_-_84716099 | 0.03 |
ENST00000560266.5
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit |
| chr6_-_73521783 | 0.03 |
ENST00000331523.7
ENST00000356303.7 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr18_+_54828406 | 0.03 |
ENST00000262094.10
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr5_+_132873660 | 0.03 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr5_-_144170607 | 0.02 |
ENST00000448443.6
ENST00000513112.5 ENST00000519064.5 ENST00000274496.10 ENST00000522203.5 |
YIPF5
|
Yip1 domain family member 5 |
| chr13_+_101489940 | 0.02 |
ENST00000376162.7
|
ITGBL1
|
integrin subunit beta like 1 |
| chr11_-_65614195 | 0.02 |
ENST00000309100.8
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr5_+_55851349 | 0.02 |
ENST00000652347.2
|
IL31RA
|
interleukin 31 receptor A |
| chr2_-_178651500 | 0.02 |
ENST00000446966.1
ENST00000426232.5 |
TTN
|
titin |
| chr11_-_2301859 | 0.02 |
ENST00000456145.2
ENST00000381153.8 |
C11orf21
|
chromosome 11 open reading frame 21 |
| chr20_-_51802433 | 0.02 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chrX_-_100874209 | 0.02 |
ENST00000372964.5
ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr1_+_50105666 | 0.02 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr5_+_55851378 | 0.01 |
ENST00000396836.6
ENST00000359040.10 |
IL31RA
|
interleukin 31 receptor A |
| chr1_-_151459169 | 0.01 |
ENST00000368863.6
ENST00000409503.5 ENST00000491586.5 ENST00000533351.5 |
POGZ
|
pogo transposable element derived with ZNF domain |
| chr2_-_202911621 | 0.01 |
ENST00000261015.5
|
WDR12
|
WD repeat domain 12 |
| chr7_+_150322639 | 0.01 |
ENST00000343855.6
|
ZBED6CL
|
ZBED6 C-terminal like |
| chr15_-_52191387 | 0.01 |
ENST00000261837.12
|
GNB5
|
G protein subunit beta 5 |
| chr13_+_45464901 | 0.01 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr1_+_50106265 | 0.01 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr4_+_122239965 | 0.01 |
ENST00000446180.5
|
KIAA1109
|
KIAA1109 |
| chr19_+_51573171 | 0.01 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chrX_-_100874351 | 0.00 |
ENST00000372966.8
|
NOX1
|
NADPH oxidase 1 |
| chr9_-_13165442 | 0.00 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr11_-_33717409 | 0.00 |
ENST00000651485.1
|
CD59
|
CD59 molecule (CD59 blood group) |
| chr1_+_107141022 | 0.00 |
ENST00000370067.5
ENST00000370068.6 |
NTNG1
|
netrin G1 |
| chr19_-_35228699 | 0.00 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.2 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.0 | GO:0097272 | ammonia homeostasis(GO:0097272) urea homeostasis(GO:0097274) |
| 0.0 | 0.1 | GO:0043418 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) homocysteine catabolic process(GO:0043418) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0050421 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |