Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for POU6F2
Z-value: 0.58
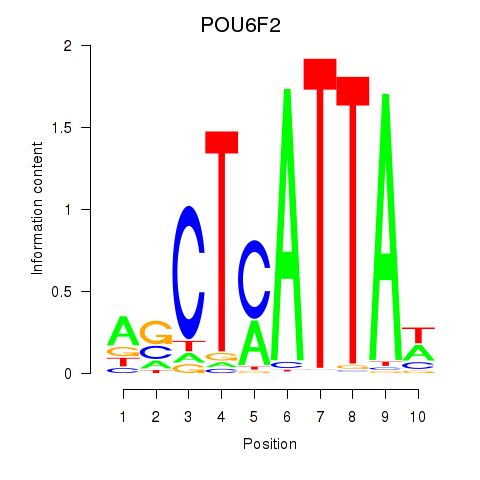
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.21 | POU6F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg38_v1_chr7_+_38977904_38978009 | 0.24 | 2.0e-01 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_127646145 | 1.50 |
ENST00000486700.2
ENST00000272644.7 |
GPR17
|
G protein-coupled receptor 17 |
| chr2_+_127645864 | 1.49 |
ENST00000544369.5
|
GPR17
|
G protein-coupled receptor 17 |
| chr12_-_30735014 | 1.39 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr6_+_151325665 | 1.23 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr1_-_182672232 | 1.11 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr19_-_14112641 | 0.90 |
ENST00000536649.5
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr3_-_33645433 | 0.89 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_+_92593112 | 0.89 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11 |
| chr5_-_140346596 | 0.87 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr12_-_86838867 | 0.85 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr2_-_68871382 | 0.80 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr12_+_15546344 | 0.73 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_-_89352395 | 0.71 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_89352487 | 0.69 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr14_+_94026314 | 0.68 |
ENST00000203664.10
ENST00000553723.1 |
OTUB2
|
OTU deubiquitinase, ubiquitin aldehyde binding 2 |
| chr3_+_159839847 | 0.67 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr1_-_208244375 | 0.59 |
ENST00000367033.4
|
PLXNA2
|
plexin A2 |
| chr14_+_32329341 | 0.57 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr6_+_63521738 | 0.56 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr8_+_22567038 | 0.51 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr6_-_87095059 | 0.48 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr8_-_10655137 | 0.46 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr12_-_16606102 | 0.46 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chr1_-_153150884 | 0.44 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr12_-_16605939 | 0.44 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr17_-_19387170 | 0.44 |
ENST00000395592.6
ENST00000299610.5 |
MFAP4
|
microfibril associated protein 4 |
| chr14_+_32329256 | 0.43 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr17_-_19386785 | 0.43 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4 |
| chr21_-_30492008 | 0.38 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr11_+_31650024 | 0.38 |
ENST00000638317.1
|
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr10_+_119207560 | 0.37 |
ENST00000392870.3
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr1_-_193059296 | 0.36 |
ENST00000367450.7
ENST00000367451.8 ENST00000367454.6 ENST00000367448.5 ENST00000367449.5 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chr5_-_157059109 | 0.34 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr14_-_68794597 | 0.34 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1 |
| chr3_+_189789734 | 0.34 |
ENST00000437221.5
ENST00000392463.6 ENST00000392461.7 ENST00000449992.5 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr6_+_130018565 | 0.33 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr6_+_25754699 | 0.33 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr1_-_193059489 | 0.32 |
ENST00000367455.8
ENST00000421683.1 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chrX_+_43656289 | 0.31 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr2_+_28395511 | 0.30 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit |
| chr14_+_39114289 | 0.29 |
ENST00000396249.7
ENST00000250379.13 ENST00000534684.7 ENST00000308317.12 ENST00000527381.2 |
GEMIN2
|
gem nuclear organelle associated protein 2 |
| chr2_-_70553440 | 0.27 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr21_-_31160904 | 0.26 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr11_-_5343524 | 0.26 |
ENST00000300773.3
|
OR51B5
|
olfactory receptor family 51 subfamily B member 5 |
| chr3_+_111999189 | 0.26 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr4_-_22443110 | 0.26 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr3_+_111998739 | 0.25 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr3_+_111998915 | 0.25 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr7_-_80512041 | 0.25 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr13_+_97960192 | 0.25 |
ENST00000496368.6
ENST00000421861.7 ENST00000357602.7 |
IPO5
|
importin 5 |
| chr3_-_71360753 | 0.24 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_101320558 | 0.24 |
ENST00000193391.8
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr3_+_189789643 | 0.23 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chrX_+_23908006 | 0.22 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr6_-_32190170 | 0.21 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr17_-_41397600 | 0.21 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr5_-_141618914 | 0.21 |
ENST00000518047.5
|
DIAPH1
|
diaphanous related formin 1 |
| chr20_+_57561103 | 0.21 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr21_-_14658812 | 0.21 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr11_-_31509569 | 0.21 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr5_+_172641241 | 0.20 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr4_+_56907876 | 0.20 |
ENST00000640168.2
ENST00000309042.12 |
REST
|
RE1 silencing transcription factor |
| chr5_-_141619049 | 0.20 |
ENST00000647433.1
ENST00000253811.10 ENST00000389057.9 ENST00000398557.8 |
DIAPH1
|
diaphanous related formin 1 |
| chr5_-_141618957 | 0.19 |
ENST00000389054.8
|
DIAPH1
|
diaphanous related formin 1 |
| chr4_-_73620629 | 0.19 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr11_-_31509588 | 0.19 |
ENST00000534812.5
ENST00000529749.5 ENST00000532287.6 ENST00000278200.5 ENST00000530023.5 ENST00000533642.1 |
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr4_-_73620391 | 0.18 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr3_+_141387616 | 0.18 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_+_108044967 | 0.17 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_13988863 | 0.17 |
ENST00000405358.8
|
ETV1
|
ETS variant transcription factor 1 |
| chr4_-_163473732 | 0.17 |
ENST00000280605.5
|
TKTL2
|
transketolase like 2 |
| chr12_-_16608183 | 0.17 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr12_-_16608073 | 0.16 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr7_-_13989658 | 0.16 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chrX_+_108045050 | 0.16 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_+_92900338 | 0.16 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr5_+_135579193 | 0.16 |
ENST00000646290.1
|
SLC25A48
|
solute carrier family 25 member 48 |
| chr12_+_53954870 | 0.16 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chr3_-_51875597 | 0.15 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5 |
| chr22_-_28306645 | 0.15 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr11_-_72112068 | 0.15 |
ENST00000537644.5
ENST00000538919.5 ENST00000539395.1 ENST00000542531.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr2_-_157444044 | 0.15 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chrX_-_155071064 | 0.15 |
ENST00000369484.8
ENST00000369476.8 |
CMC4
MTCP1
|
C-X9-C motif containing 4 mature T cell proliferation 1 |
| chr17_-_41315706 | 0.14 |
ENST00000334202.5
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr6_+_29306626 | 0.14 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr11_-_72112750 | 0.14 |
ENST00000545680.5
ENST00000543587.5 ENST00000538393.5 ENST00000535234.5 ENST00000227618.8 ENST00000535503.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr11_-_72112669 | 0.14 |
ENST00000545944.5
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr6_-_89217339 | 0.14 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr17_-_41424583 | 0.14 |
ENST00000225550.4
|
KRT37
|
keratin 37 |
| chr4_+_109827963 | 0.13 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr21_-_40847149 | 0.13 |
ENST00000400454.6
|
DSCAM
|
DS cell adhesion molecule |
| chrX_+_69616067 | 0.13 |
ENST00000338901.4
ENST00000525810.5 ENST00000527388.5 ENST00000374553.6 ENST00000374552.9 ENST00000524573.5 |
EDA
|
ectodysplasin A |
| chr3_-_127822455 | 0.12 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr8_+_26293112 | 0.12 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr10_-_92497727 | 0.12 |
ENST00000496903.5
ENST00000678824.1 ENST00000371581.9 |
IDE
|
insulin degrading enzyme |
| chr13_+_57631735 | 0.11 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr6_+_121437378 | 0.11 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr3_+_111999326 | 0.10 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_+_112860912 | 0.10 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr7_+_116222804 | 0.10 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr15_+_92900189 | 0.10 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr1_+_50106265 | 0.10 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr12_-_52652207 | 0.10 |
ENST00000309680.4
|
KRT2
|
keratin 2 |
| chrX_-_23907887 | 0.10 |
ENST00000379226.9
|
APOO
|
apolipoprotein O |
| chr4_+_112861053 | 0.10 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr15_+_58410543 | 0.09 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr4_+_112860981 | 0.09 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr6_-_155314444 | 0.09 |
ENST00000367166.5
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr10_+_35195843 | 0.09 |
ENST00000488741.5
ENST00000474931.5 ENST00000468236.5 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr11_-_36598221 | 0.09 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr17_-_42185452 | 0.09 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr12_-_52553139 | 0.08 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr2_-_2326378 | 0.08 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr18_+_63702958 | 0.08 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr1_+_175067831 | 0.08 |
ENST00000239462.9
|
TNN
|
tenascin N |
| chr13_-_44161257 | 0.08 |
ENST00000400419.2
|
SMIM2
|
small integral membrane protein 2 |
| chr7_+_148590760 | 0.07 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr13_-_103066411 | 0.07 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr9_+_106863121 | 0.07 |
ENST00000472574.1
ENST00000277225.10 |
ZNF462
|
zinc finger protein 462 |
| chr15_+_74788542 | 0.07 |
ENST00000567571.5
|
CSK
|
C-terminal Src kinase |
| chr4_+_40196907 | 0.07 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chrX_+_30235894 | 0.07 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr18_-_13915531 | 0.06 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor |
| chr8_+_76681208 | 0.06 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr19_-_7633713 | 0.06 |
ENST00000311069.6
|
PCP2
|
Purkinje cell protein 2 |
| chr9_-_114806031 | 0.06 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15 |
| chr6_+_158017048 | 0.06 |
ENST00000638626.1
|
SYNJ2
|
synaptojanin 2 |
| chr19_+_9178979 | 0.06 |
ENST00000642043.1
ENST00000641288.2 |
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chrX_-_21658324 | 0.06 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr4_+_40197023 | 0.06 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr13_+_45464901 | 0.06 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr11_+_107591077 | 0.05 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr1_+_50105666 | 0.05 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr16_-_18926408 | 0.05 |
ENST00000446231.7
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr3_-_57165332 | 0.05 |
ENST00000296318.12
|
IL17RD
|
interleukin 17 receptor D |
| chr10_-_101229449 | 0.05 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr21_-_34526815 | 0.05 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr12_+_120694167 | 0.04 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr3_+_77039836 | 0.04 |
ENST00000461745.5
|
ROBO2
|
roundabout guidance receptor 2 |
| chr2_-_2324323 | 0.04 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr7_-_19145306 | 0.04 |
ENST00000275461.3
|
FERD3L
|
Fer3 like bHLH transcription factor |
| chr10_+_18260715 | 0.04 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr21_-_30881572 | 0.04 |
ENST00000332378.6
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr20_-_51802433 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr19_+_38374557 | 0.04 |
ENST00000215071.9
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr19_+_38374549 | 0.03 |
ENST00000620216.4
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr4_+_87650277 | 0.03 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr1_+_45913647 | 0.03 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr10_+_112950586 | 0.03 |
ENST00000355717.9
ENST00000352065.10 ENST00000369395.6 ENST00000545257.6 |
TCF7L2
|
transcription factor 7 like 2 |
| chrX_-_18672101 | 0.03 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr19_-_7632971 | 0.03 |
ENST00000598935.5
|
PCP2
|
Purkinje cell protein 2 |
| chr9_-_34729482 | 0.03 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A |
| chr19_-_49423441 | 0.03 |
ENST00000270631.2
|
PTH2
|
parathyroid hormone 2 |
| chr2_+_162318884 | 0.03 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr1_-_47190013 | 0.03 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr17_+_39628496 | 0.02 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr1_-_182671853 | 0.02 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr5_+_127649018 | 0.02 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr1_-_21279520 | 0.02 |
ENST00000357071.8
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_158426237 | 0.02 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr3_+_141386862 | 0.02 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_112950240 | 0.01 |
ENST00000627217.3
ENST00000534894.5 ENST00000538897.5 ENST00000536810.5 ENST00000355995.8 ENST00000542695.5 ENST00000543371.5 |
TCF7L2
|
transcription factor 7 like 2 |
| chr1_-_182671902 | 0.01 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8 |
| chr7_-_126533850 | 0.01 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr17_+_7407838 | 0.01 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr20_+_43558968 | 0.00 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr8_-_42501224 | 0.00 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr6_+_50818857 | 0.00 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr10_+_112950452 | 0.00 |
ENST00000369397.8
|
TCF7L2
|
transcription factor 7 like 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.7 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 3.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 1.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.8 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.3 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.9 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.2 | GO:2000797 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.1 | 0.3 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.7 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 1.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.9 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 2.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 3.0 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |