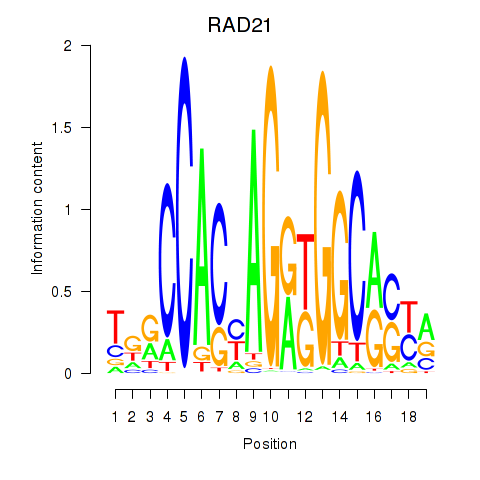
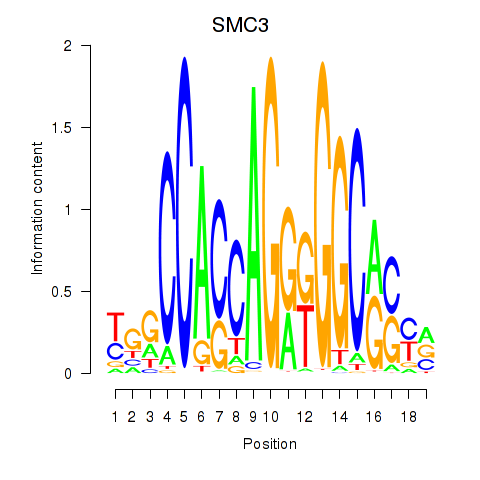
|
chr5_+_149141817
Show fit
|
4.09 |
ENST00000504238.5
|
ABLIM3
|
actin binding LIM protein family member 3
|
|
chr5_+_149141483
Show fit
|
3.97 |
ENST00000326685.11
ENST00000309868.12
|
ABLIM3
|
actin binding LIM protein family member 3
|
|
chr5_+_149141573
Show fit
|
3.94 |
ENST00000506113.5
|
ABLIM3
|
actin binding LIM protein family member 3
|
|
chr4_+_74933095
Show fit
|
2.39 |
ENST00000513238.5
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr4_+_74933108
Show fit
|
2.36 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr4_+_7192519
Show fit
|
2.18 |
ENST00000507866.6
|
SORCS2
|
sortilin related VPS10 domain containing receptor 2
|
|
chr19_-_1513003
Show fit
|
1.46 |
ENST00000330475.9
ENST00000586272.5
ENST00000590562.5
|
ADAMTSL5
|
ADAMTS like 5
|
|
chr7_+_90154442
Show fit
|
1.43 |
ENST00000297205.7
|
STEAP1
|
STEAP family member 1
|
|
chr17_-_74361860
Show fit
|
1.36 |
ENST00000375366.4
|
BTBD17
|
BTB domain containing 17
|
|
chr15_+_45430579
Show fit
|
1.35 |
ENST00000558435.5
ENST00000344300.3
ENST00000396650.7
|
C15orf48
|
chromosome 15 open reading frame 48
|
|
chr19_-_18941117
Show fit
|
1.27 |
ENST00000600077.5
|
HOMER3
|
homer scaffold protein 3
|
|
chr11_-_17389323
Show fit
|
1.24 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly rectifying channel subfamily J member 11
|
|
chr3_-_184017863
Show fit
|
1.23 |
ENST00000427120.6
ENST00000334444.11
ENST00000392579.6
ENST00000382494.6
ENST00000265586.10
ENST00000446941.2
|
ABCC5
|
ATP binding cassette subfamily C member 5
|
|
chr15_+_73873604
Show fit
|
1.19 |
ENST00000535547.6
ENST00000562056.1
|
TBC1D21
|
TBC1 domain family member 21
|
|
chr12_-_51269949
Show fit
|
1.18 |
ENST00000603864.5
ENST00000605426.5
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr14_-_93115812
Show fit
|
1.10 |
ENST00000553452.5
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase
|
|
chr19_-_18941184
Show fit
|
1.07 |
ENST00000594794.5
ENST00000392351.8
ENST00000596482.5
|
HOMER3
|
homer scaffold protein 3
|
|
chr16_+_30748241
Show fit
|
1.02 |
ENST00000565924.5
ENST00000424889.7
|
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2
|
|
chr6_+_137867241
Show fit
|
0.99 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr5_-_77492309
Show fit
|
0.99 |
ENST00000296679.9
ENST00000507029.5
|
WDR41
|
WD repeat domain 41
|
|
chr11_-_117232033
Show fit
|
0.97 |
ENST00000524507.6
ENST00000320934.8
ENST00000532301.5
ENST00000676339.1
ENST00000540028.5
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr15_-_65422894
Show fit
|
0.96 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4
|
|
chr19_-_4400418
Show fit
|
0.94 |
ENST00000598564.5
ENST00000417295.6
ENST00000269886.7
|
SH3GL1
|
SH3 domain containing GRB2 like 1, endophilin A2
|
|
chr6_+_137867414
Show fit
|
0.94 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr19_-_48835823
Show fit
|
0.92 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid 17-beta dehydrogenase 14
|
|
chr12_-_51270175
Show fit
|
0.91 |
ENST00000604188.1
ENST00000398453.7
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr19_-_15479469
Show fit
|
0.89 |
ENST00000292609.8
ENST00000340880.5
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr14_+_21841182
Show fit
|
0.89 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1
|
|
chr20_-_31723491
Show fit
|
0.88 |
ENST00000676582.1
ENST00000422920.2
|
BCL2L1
|
BCL2 like 1
|
|
chr5_-_140346596
Show fit
|
0.86 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor
|
|
chr17_+_76868410
Show fit
|
0.86 |
ENST00000301618.8
|
MGAT5B
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase B
|
|
chr3_+_5122276
Show fit
|
0.85 |
ENST00000256496.8
ENST00000419534.2
|
ARL8B
|
ADP ribosylation factor like GTPase 8B
|
|
chr8_-_27992624
Show fit
|
0.84 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5
|
|
chr10_-_29735787
Show fit
|
0.84 |
ENST00000375400.7
|
SVIL
|
supervillin
|
|
chr3_+_126983035
Show fit
|
0.83 |
ENST00000393409.3
|
PLXNA1
|
plexin A1
|
|
chr20_-_31723902
Show fit
|
0.83 |
ENST00000676942.1
ENST00000450273.2
ENST00000678563.1
ENST00000456404.6
ENST00000420488.6
ENST00000439267.2
|
BCL2L1
|
BCL2 like 1
|
|
chr19_-_45815303
Show fit
|
0.82 |
ENST00000221538.8
|
RSPH6A
|
radial spoke head 6 homolog A
|
|
chr17_+_78232006
Show fit
|
0.82 |
ENST00000550981.7
ENST00000591033.2
|
TMEM235
|
transmembrane protein 235
|
|
chr11_+_118956289
Show fit
|
0.80 |
ENST00000264031.3
|
UPK2
|
uroplakin 2
|
|
chr5_-_137499293
Show fit
|
0.80 |
ENST00000510689.5
ENST00000394945.6
|
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1
|
|
chr1_+_47438036
Show fit
|
0.80 |
ENST00000334793.6
|
FOXD2
|
forkhead box D2
|
|
chr1_+_37474572
Show fit
|
0.80 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr1_-_159945596
Show fit
|
0.79 |
ENST00000361509.7
ENST00000611023.1
ENST00000368094.6
|
IGSF9
|
immunoglobulin superfamily member 9
|
|
chr19_-_55140922
Show fit
|
0.78 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type
|
|
chr19_-_45815228
Show fit
|
0.78 |
ENST00000597055.1
|
RSPH6A
|
radial spoke head 6 homolog A
|
|
chr8_+_22165140
Show fit
|
0.77 |
ENST00000397814.7
ENST00000354870.5
|
BMP1
|
bone morphogenetic protein 1
|
|
chr17_+_76868396
Show fit
|
0.76 |
ENST00000569840.7
|
MGAT5B
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase B
|
|
chr17_+_47733228
Show fit
|
0.74 |
ENST00000177694.2
|
TBX21
|
T-box transcription factor 21
|
|
chr15_-_38564635
Show fit
|
0.72 |
ENST00000450598.6
ENST00000559830.5
ENST00000558164.5
ENST00000539159.5
ENST00000310803.10
|
RASGRP1
|
RAS guanyl releasing protein 1
|
|
chr19_-_1513189
Show fit
|
0.71 |
ENST00000395467.6
|
ADAMTSL5
|
ADAMTS like 5
|
|
chr16_+_66604782
Show fit
|
0.71 |
ENST00000565003.5
|
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3
|
|
chr5_-_77492199
Show fit
|
0.70 |
ENST00000515253.5
ENST00000507654.5
ENST00000514559.5
ENST00000511791.1
|
WDR41
|
WD repeat domain 41
|
|
chr19_+_57830288
Show fit
|
0.69 |
ENST00000442832.8
ENST00000594901.2
|
ZNF587B
|
zinc finger protein 587B
|
|
chr16_-_88785210
Show fit
|
0.69 |
ENST00000301015.14
|
PIEZO1
|
piezo type mechanosensitive ion channel component 1
|
|
chr9_+_137423350
Show fit
|
0.69 |
ENST00000341349.6
ENST00000683555.1
ENST00000392815.2
ENST00000683683.1
|
NOXA1
|
NADPH oxidase activator 1
|
|
chr19_+_47713412
Show fit
|
0.68 |
ENST00000538399.1
ENST00000263277.8
|
EHD2
|
EH domain containing 2
|
|
chr3_+_141876124
Show fit
|
0.67 |
ENST00000475483.5
|
ATP1B3
|
ATPase Na+/K+ transporting subunit beta 3
|
|
chr22_-_21938557
Show fit
|
0.67 |
ENST00000424647.1
ENST00000407142.5
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F
|
|
chr5_+_177087412
Show fit
|
0.66 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr14_+_21797272
Show fit
|
0.66 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1
|
|
chr3_+_58306236
Show fit
|
0.66 |
ENST00000295959.10
ENST00000445193.7
ENST00000466547.1
ENST00000475412.5
ENST00000474660.5
ENST00000477305.5
ENST00000481972.5
|
RPP14
HTD2
|
ribonuclease P/MRP subunit p14
hydroxyacyl-thioester dehydratase type 2
|
|
chr19_-_55141889
Show fit
|
0.65 |
ENST00000593194.5
|
TNNT1
|
troponin T1, slow skeletal type
|
|
chr20_+_31514410
Show fit
|
0.65 |
ENST00000335574.10
ENST00000340852.9
ENST00000398174.9
ENST00000466766.2
ENST00000498035.5
ENST00000344042.5
|
HM13
|
histocompatibility minor 13
|
|
chr7_-_38349972
Show fit
|
0.65 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr10_+_97572771
Show fit
|
0.65 |
ENST00000370655.6
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2
|
|
chr3_+_58306505
Show fit
|
0.64 |
ENST00000461393.7
|
HTD2
|
hydroxyacyl-thioester dehydratase type 2
|
|
chr1_+_92029971
Show fit
|
0.64 |
ENST00000370383.5
|
EPHX4
|
epoxide hydrolase 4
|
|
chr10_+_97572493
Show fit
|
0.63 |
ENST00000307518.9
ENST00000298808.9
|
ANKRD2
|
ankyrin repeat domain 2
|
|
chr19_-_19643547
Show fit
|
0.63 |
ENST00000587238.5
|
GMIP
|
GEM interacting protein
|
|
chr19_+_10013468
Show fit
|
0.62 |
ENST00000591589.3
|
RDH8
|
retinol dehydrogenase 8
|
|
chr1_+_17755307
Show fit
|
0.61 |
ENST00000375406.2
|
ACTL8
|
actin like 8
|
|
chr14_+_22040576
Show fit
|
0.61 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20
|
|
chr19_-_41353904
Show fit
|
0.61 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1
|
|
chr2_-_165953750
Show fit
|
0.61 |
ENST00000243344.8
ENST00000679799.1
ENST00000679840.1
ENST00000681606.1
ENST00000680448.1
|
TTC21B
|
tetratricopeptide repeat domain 21B
|
|
chr3_-_128493173
Show fit
|
0.61 |
ENST00000498200.1
ENST00000341105.7
|
GATA2
|
GATA binding protein 2
|
|
chr14_+_21887848
Show fit
|
0.60 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2
|
|
chr1_-_74733253
Show fit
|
0.60 |
ENST00000417775.5
|
CRYZ
|
crystallin zeta
|
|
chr17_-_42681840
Show fit
|
0.59 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10
|
|
chr15_-_78811415
Show fit
|
0.59 |
ENST00000388820.5
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif 7
|
|
chr22_+_31082860
Show fit
|
0.59 |
ENST00000619644.4
|
SMTN
|
smoothelin
|
|
chr16_-_30021288
Show fit
|
0.59 |
ENST00000574405.5
|
DOC2A
|
double C2 domain alpha
|
|
chr8_-_144326842
Show fit
|
0.58 |
ENST00000528718.6
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr11_-_66336396
Show fit
|
0.58 |
ENST00000627248.1
ENST00000311320.9
|
RIN1
|
Ras and Rab interactor 1
|
|
chr15_-_34210011
Show fit
|
0.58 |
ENST00000557877.5
|
KATNBL1
|
katanin regulatory subunit B1 like 1
|
|
chr17_-_74923222
Show fit
|
0.58 |
ENST00000614341.5
|
USH1G
|
USH1 protein network component sans
|
|
chr17_-_45132505
Show fit
|
0.58 |
ENST00000619929.5
|
PLCD3
|
phospholipase C delta 3
|
|
chr14_+_51651858
Show fit
|
0.57 |
ENST00000395718.6
|
FRMD6
|
FERM domain containing 6
|
|
chr14_+_21852457
Show fit
|
0.57 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3
|
|
chr11_+_60924452
Show fit
|
0.57 |
ENST00000453848.7
ENST00000544065.5
ENST00000005286.8
|
TMEM132A
|
transmembrane protein 132A
|
|
chr7_+_112480853
Show fit
|
0.57 |
ENST00000439068.6
ENST00000312849.4
|
LSMEM1
|
leucine rich single-pass membrane protein 1
|
|
chr10_-_48524236
Show fit
|
0.57 |
ENST00000374170.5
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr1_-_156705764
Show fit
|
0.56 |
ENST00000621784.4
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr1_-_156705742
Show fit
|
0.56 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr2_-_24084292
Show fit
|
0.56 |
ENST00000413037.1
ENST00000407482.5
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr20_+_45363454
Show fit
|
0.56 |
ENST00000453003.1
|
SYS1
|
SYS1 golgi trafficking protein
|
|
chr8_+_22589240
Show fit
|
0.55 |
ENST00000450780.6
ENST00000430850.6
ENST00000447849.2
ENST00000614502.4
ENST00000443561.3
|
ENSG00000248235.6
PDLIM2
|
novel protein
PDZ and LIM domain 2
|
|
chr1_-_211133945
Show fit
|
0.54 |
ENST00000640044.1
ENST00000640566.1
|
KCNH1
|
potassium voltage-gated channel subfamily H member 1
|
|
chr14_+_51651901
Show fit
|
0.54 |
ENST00000344768.10
|
FRMD6
|
FERM domain containing 6
|
|
chr7_+_1086800
Show fit
|
0.54 |
ENST00000413368.5
ENST00000397092.5
ENST00000297469.3
|
GPER1
|
G protein-coupled estrogen receptor 1
|
|
chr10_-_88583304
Show fit
|
0.54 |
ENST00000331772.9
|
RNLS
|
renalase, FAD dependent amine oxidase
|
|
chr1_+_203682734
Show fit
|
0.53 |
ENST00000341360.6
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4
|
|
chr18_+_46174055
Show fit
|
0.52 |
ENST00000615553.1
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr17_-_47831509
Show fit
|
0.51 |
ENST00000414011.1
ENST00000351111.7
|
MRPL10
|
mitochondrial ribosomal protein L10
|
|
chr2_+_227813834
Show fit
|
0.51 |
ENST00000358813.5
ENST00000409189.7
|
CCL20
|
C-C motif chemokine ligand 20
|
|
chr6_+_35342614
Show fit
|
0.51 |
ENST00000337400.6
ENST00000311565.4
|
PPARD
|
peroxisome proliferator activated receptor delta
|
|
chr17_+_74203582
Show fit
|
0.50 |
ENST00000439590.6
ENST00000311111.11
ENST00000584577.5
ENST00000534490.5
ENST00000528433.2
ENST00000533498.1
|
RPL38
|
ribosomal protein L38
|
|
chr6_-_11044275
Show fit
|
0.50 |
ENST00000354666.4
|
ELOVL2
|
ELOVL fatty acid elongase 2
|
|
chr16_+_30748378
Show fit
|
0.50 |
ENST00000563588.6
ENST00000328273.11
|
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2
|
|
chr10_+_102394488
Show fit
|
0.50 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr12_-_95116967
Show fit
|
0.49 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr16_+_67645100
Show fit
|
0.49 |
ENST00000334583.11
|
CARMIL2
|
capping protein regulator and myosin 1 linker 2
|
|
chr9_+_136844415
Show fit
|
0.48 |
ENST00000436881.2
|
AJM1
|
apical junction component 1 homolog
|
|
chr16_-_70680025
Show fit
|
0.48 |
ENST00000576338.1
|
MTSS2
|
MTSS I-BAR domain containing 2
|
|
chr1_-_145910031
Show fit
|
0.47 |
ENST00000369304.8
|
ITGA10
|
integrin subunit alpha 10
|
|
chr2_-_218292496
Show fit
|
0.47 |
ENST00000258412.8
ENST00000445635.5
ENST00000413976.1
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1
|
|
chr7_+_73667824
Show fit
|
0.46 |
ENST00000324941.5
ENST00000451519.1
|
VPS37D
|
VPS37D subunit of ESCRT-I
|
|
chr9_-_109321041
Show fit
|
0.46 |
ENST00000374566.8
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr8_+_28622729
Show fit
|
0.46 |
ENST00000523149.5
|
EXTL3
|
exostosin like glycosyltransferase 3
|
|
chr9_-_21141832
Show fit
|
0.46 |
ENST00000380229.4
|
IFNW1
|
interferon omega 1
|
|
chr12_-_120265719
Show fit
|
0.46 |
ENST00000637617.2
ENST00000267257.11
ENST00000228307.11
ENST00000424649.6
|
PXN
|
paxillin
|
|
chr22_+_22594528
Show fit
|
0.45 |
ENST00000390303.3
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional)
|
|
chr1_-_145910066
Show fit
|
0.45 |
ENST00000539363.2
|
ITGA10
|
integrin subunit alpha 10
|
|
chr3_+_49171590
Show fit
|
0.45 |
ENST00000332780.4
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr22_-_22559229
Show fit
|
0.44 |
ENST00000405655.8
ENST00000406503.1
ENST00000439106.5
ENST00000402697.5
ENST00000543184.5
ENST00000398743.6
|
PRAME
|
PRAME nuclear receptor transcriptional regulator
|
|
chr12_+_112418976
Show fit
|
0.44 |
ENST00000635625.1
|
PTPN11
|
protein tyrosine phosphatase non-receptor type 11
|
|
chr18_-_50819982
Show fit
|
0.44 |
ENST00000398439.8
ENST00000431965.6
ENST00000436348.6
|
MRO
|
maestro
|
|
chr16_+_67645166
Show fit
|
0.43 |
ENST00000545661.5
|
CARMIL2
|
capping protein regulator and myosin 1 linker 2
|
|
chr10_-_15719885
Show fit
|
0.43 |
ENST00000378076.4
|
ITGA8
|
integrin subunit alpha 8
|
|
chr18_+_596973
Show fit
|
0.43 |
ENST00000579912.5
ENST00000400606.6
ENST00000540035.5
|
CLUL1
|
clusterin like 1
|
|
chr19_-_19643597
Show fit
|
0.42 |
ENST00000587205.1
ENST00000203556.9
|
GMIP
|
GEM interacting protein
|
|
chr2_-_24084822
Show fit
|
0.42 |
ENST00000238721.9
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr16_-_11256192
Show fit
|
0.42 |
ENST00000644787.1
ENST00000332029.4
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr17_+_45135640
Show fit
|
0.42 |
ENST00000586346.5
ENST00000321854.13
ENST00000398322.7
ENST00000592162.5
ENST00000376955.8
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr16_+_66604696
Show fit
|
0.42 |
ENST00000567572.6
ENST00000564060.5
ENST00000565922.1
|
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3
|
|
chr3_-_197029775
Show fit
|
0.41 |
ENST00000439320.1
ENST00000296351.8
ENST00000296350.10
|
MELTF
|
melanotransferrin
|
|
chr11_+_61752603
Show fit
|
0.41 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor
|
|
chr17_+_7717727
Show fit
|
0.41 |
ENST00000570791.5
|
DNAH2
|
dynein axonemal heavy chain 2
|
|
chr6_+_35342535
Show fit
|
0.40 |
ENST00000360694.8
ENST00000418635.6
ENST00000448077.6
|
PPARD
|
peroxisome proliferator activated receptor delta
|
|
chr8_-_144326908
Show fit
|
0.40 |
ENST00000332324.5
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr9_+_128203371
Show fit
|
0.40 |
ENST00000475805.5
ENST00000372923.8
ENST00000341179.11
ENST00000393594.7
|
DNM1
|
dynamin 1
|
|
chr17_-_44363839
Show fit
|
0.39 |
ENST00000293443.12
|
FAM171A2
|
family with sequence similarity 171 member A2
|
|
chr19_-_35135180
Show fit
|
0.39 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr16_+_23557714
Show fit
|
0.38 |
ENST00000567212.5
ENST00000567264.1
ENST00000395878.8
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chr22_-_21952827
Show fit
|
0.38 |
ENST00000397495.8
ENST00000263212.10
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F
|
|
chr3_-_136752361
Show fit
|
0.38 |
ENST00000480733.1
ENST00000629124.2
ENST00000383202.7
ENST00000236698.9
ENST00000434713.6
|
STAG1
|
stromal antigen 1
|
|
chr17_-_8079903
Show fit
|
0.38 |
ENST00000649809.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type
|
|
chr1_+_21509415
Show fit
|
0.37 |
ENST00000374840.8
ENST00000539907.5
ENST00000540617.5
|
ALPL
|
alkaline phosphatase, biomineralization associated
|
|
chr17_-_7614824
Show fit
|
0.37 |
ENST00000571597.1
ENST00000250113.12
|
FXR2
|
FMR1 autosomal homolog 2
|
|
chr3_-_49719668
Show fit
|
0.37 |
ENST00000320431.8
|
AMIGO3
|
adhesion molecule with Ig like domain 3
|
|
chr10_-_75109172
Show fit
|
0.37 |
ENST00000372700.7
ENST00000473072.2
ENST00000491677.6
ENST00000372702.7
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr16_+_16232516
Show fit
|
0.37 |
ENST00000399336.9
ENST00000678538.1
ENST00000678192.1
ENST00000677777.1
ENST00000677260.1
ENST00000676550.1
ENST00000263012.10
|
NOMO3
|
NODAL modulator 3
|
|
chr19_+_1941118
Show fit
|
0.37 |
ENST00000255641.13
|
CSNK1G2
|
casein kinase 1 gamma 2
|
|
chr1_-_151716052
Show fit
|
0.36 |
ENST00000290585.8
|
CELF3
|
CUGBP Elav-like family member 3
|
|
chr20_+_31440626
Show fit
|
0.36 |
ENST00000376309.4
|
DEFB123
|
defensin beta 123
|
|
chr14_-_64823148
Show fit
|
0.36 |
ENST00000389722.7
|
SPTB
|
spectrin beta, erythrocytic
|
|
chr1_+_145845608
Show fit
|
0.36 |
ENST00000334513.6
|
NUDT17
|
nudix hydrolase 17
|
|
chr11_+_117232625
Show fit
|
0.36 |
ENST00000534428.5
ENST00000300650.9
|
RNF214
|
ring finger protein 214
|
|
chr6_+_31137646
Show fit
|
0.36 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr19_+_55654115
Show fit
|
0.35 |
ENST00000450554.6
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr17_+_2055094
Show fit
|
0.35 |
ENST00000399849.4
ENST00000619757.5
|
HIC1
|
HIC ZBTB transcriptional repressor 1
|
|
chr15_-_44195210
Show fit
|
0.35 |
ENST00000402883.5
ENST00000417257.6
|
FRMD5
|
FERM domain containing 5
|
|
chr10_-_90857680
Show fit
|
0.35 |
ENST00000277874.10
ENST00000371719.2
|
HTR7
|
5-hydroxytryptamine receptor 7
|
|
chr7_-_22500152
Show fit
|
0.34 |
ENST00000406890.6
ENST00000678116.1
ENST00000424363.5
|
STEAP1B
|
STEAP family member 1B
|
|
chr18_-_50819764
Show fit
|
0.34 |
ENST00000592966.5
|
MRO
|
maestro
|
|
chr10_-_75235917
Show fit
|
0.34 |
ENST00000469299.1
ENST00000372538.8
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr10_-_90857983
Show fit
|
0.34 |
ENST00000336152.8
|
HTR7
|
5-hydroxytryptamine receptor 7
|
|
chr10_-_101843920
Show fit
|
0.33 |
ENST00000358038.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2
|
|
chr10_-_75109085
Show fit
|
0.33 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13
|
|
chrX_-_132219473
Show fit
|
0.33 |
ENST00000620646.4
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr11_+_85855377
Show fit
|
0.33 |
ENST00000342404.8
|
CCDC83
|
coiled-coil domain containing 83
|
|
chr2_-_231530427
Show fit
|
0.33 |
ENST00000305141.5
|
NMUR1
|
neuromedin U receptor 1
|
|
chr9_+_126860625
Show fit
|
0.33 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr11_+_61680373
Show fit
|
0.33 |
ENST00000257215.10
|
DAGLA
|
diacylglycerol lipase alpha
|
|
chr13_+_87671354
Show fit
|
0.33 |
ENST00000683689.1
|
SLITRK5
|
SLIT and NTRK like family member 5
|
|
chrX_-_132219439
Show fit
|
0.32 |
ENST00000370874.2
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr19_-_4558417
Show fit
|
0.32 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B
|
|
chr5_+_140821598
Show fit
|
0.32 |
ENST00000614258.1
ENST00000529859.2
ENST00000529619.5
|
PCDHA5
|
protocadherin alpha 5
|
|
chr16_+_3654683
Show fit
|
0.32 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1
|
|
chr3_+_134795277
Show fit
|
0.30 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1
|
|
chr11_+_126269110
Show fit
|
0.30 |
ENST00000263578.10
ENST00000532125.1
|
FOXRED1
|
FAD dependent oxidoreductase domain containing 1
|
|
chr10_+_100999287
Show fit
|
0.30 |
ENST00000370220.1
|
LZTS2
|
leucine zipper tumor suppressor 2
|
|
chr11_+_119206298
Show fit
|
0.30 |
ENST00000634586.1
ENST00000634840.1
ENST00000264033.6
ENST00000637974.1
|
CBL
|
Cbl proto-oncogene
|
|
chr3_+_184561768
Show fit
|
0.30 |
ENST00000330394.3
|
EPHB3
|
EPH receptor B3
|
|
chr17_+_16690223
Show fit
|
0.29 |
ENST00000340621.9
ENST00000399273.5
ENST00000456009.4
ENST00000360524.12
|
CCDC144A
|
coiled-coil domain containing 144A
|
|
chr15_+_31978056
Show fit
|
0.29 |
ENST00000637183.1
|
CHRNA7
|
cholinergic receptor nicotinic alpha 7 subunit
|
|
chr19_-_1848452
Show fit
|
0.29 |
ENST00000170168.9
|
REXO1
|
RNA exonuclease 1 homolog
|
|
chr3_-_138947095
Show fit
|
0.29 |
ENST00000648323.1
|
FOXL2
|
forkhead box L2
|
|
chr12_+_50504970
Show fit
|
0.29 |
ENST00000301180.10
|
DIP2B
|
disco interacting protein 2 homolog B
|
|
chr22_+_23070496
Show fit
|
0.29 |
ENST00000615612.2
|
GNAZ
|
G protein subunit alpha z
|
|
chr22_-_22559073
Show fit
|
0.29 |
ENST00000420709.5
ENST00000398741.5
|
PRAME
|
PRAME nuclear receptor transcriptional regulator
|
|
chr3_+_39809602
Show fit
|
0.28 |
ENST00000302541.11
ENST00000396217.7
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr4_-_170090153
Show fit
|
0.28 |
ENST00000509167.5
ENST00000353187.6
ENST00000507375.5
ENST00000515480.5
|
AADAT
|
aminoadipate aminotransferase
|
|
chr7_-_5529949
Show fit
|
0.28 |
ENST00000642480.2
ENST00000417101.2
|
ACTB
|
actin beta
|
|
chr1_-_34859717
Show fit
|
0.28 |
ENST00000423898.1
ENST00000521580.3
ENST00000456842.1
|
SMIM12
|
small integral membrane protein 12
|
|
chr20_+_45363114
Show fit
|
0.27 |
ENST00000426004.5
ENST00000243918.10
|
SYS1
|
SYS1 golgi trafficking protein
|
|
chr9_+_131009141
Show fit
|
0.27 |
ENST00000361069.9
|
LAMC3
|
laminin subunit gamma 3
|
|
chr17_+_43483949
Show fit
|
0.27 |
ENST00000540306.5
ENST00000262415.8
|
DHX8
|
DEAH-box helicase 8
|
|
chr9_+_128203397
Show fit
|
0.27 |
ENST00000628346.2
ENST00000486160.3
ENST00000627061.2
ENST00000627543.2
ENST00000634267.2
|
DNM1
|
dynamin 1
|
|
chr13_-_52450590
Show fit
|
0.27 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog
|
|
chr2_+_69013414
Show fit
|
0.26 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|