Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for RFX7_RFX4_RFX1
Z-value: 10.66
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
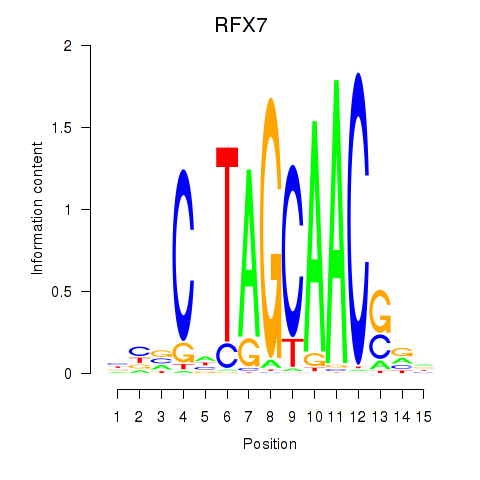
RFX7
|
ENSG00000181827.16 | RFX7 |
|
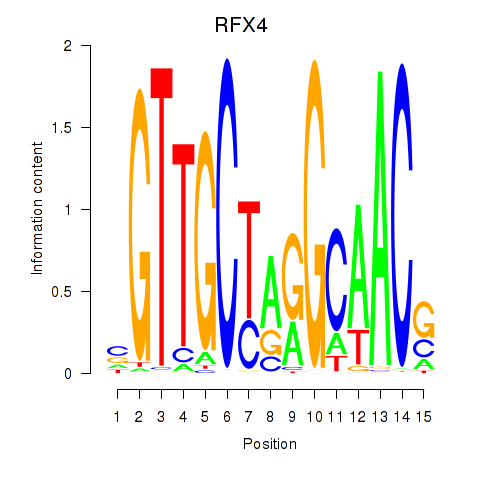
RFX4
|
ENSG00000111783.13 | RFX4 |
|
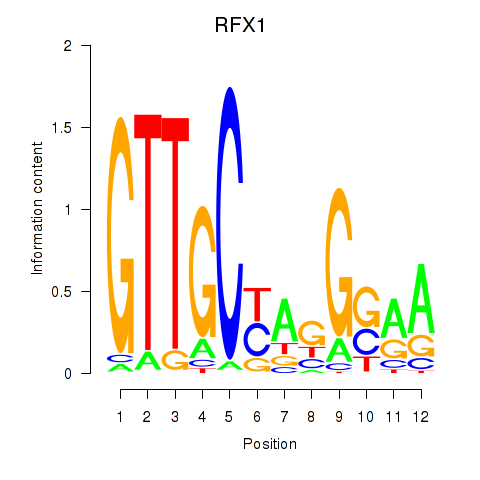
RFX1
|
ENSG00000132005.9 | RFX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg38_v1_chr15_-_56245074_56245088, hg38_v1_chr15_-_56243829_56243891 | -0.50 | 4.8e-03 | Click! |
| RFX4 | hg38_v1_chr12_+_106684696_106684755, hg38_v1_chr12_+_106582996_106583010 | -0.30 | 1.0e-01 | Click! |
| RFX1 | hg38_v1_chr19_-_14006262_14006330 | -0.25 | 1.9e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 41.3 | 165.1 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 23.8 | 71.5 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 21.7 | 65.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 19.4 | 97.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 16.1 | 64.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 14.2 | 127.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 12.0 | 240.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 9.6 | 57.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 9.4 | 375.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 8.3 | 41.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 8.3 | 99.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 7.7 | 23.2 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 7.5 | 22.5 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 6.8 | 115.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 6.4 | 121.3 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 6.3 | 112.9 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 6.1 | 18.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 5.9 | 5.9 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 5.9 | 17.6 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 5.9 | 52.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 5.0 | 25.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 5.0 | 15.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 4.9 | 4.9 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 4.7 | 14.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 4.3 | 298.1 | GO:0042073 | intraciliary transport(GO:0042073) |
| 4.1 | 32.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 4.1 | 16.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 4.0 | 63.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 3.9 | 19.7 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 3.7 | 14.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 3.7 | 69.6 | GO:0003341 | cilium movement(GO:0003341) |
| 3.3 | 10.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 3.3 | 59.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 3.1 | 21.8 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 3.0 | 36.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 2.9 | 8.7 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 2.9 | 8.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 2.6 | 31.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 2.5 | 2.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 2.4 | 7.2 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 2.3 | 6.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 1.6 | 4.9 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 1.5 | 4.5 | GO:1902214 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 1.5 | 31.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.4 | 4.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.4 | 2.8 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 1.4 | 4.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 1.3 | 1.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.2 | 11.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.1 | 9.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 1.1 | 149.3 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 1.0 | 14.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 1.0 | 141.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 1.0 | 96.9 | GO:0044782 | cilium organization(GO:0044782) |
| 0.9 | 2.8 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.9 | 24.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.9 | 8.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.9 | 4.4 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.8 | 11.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.8 | 6.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.8 | 6.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.7 | 2.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) regulation of amacrine cell differentiation(GO:1902869) |
| 0.7 | 5.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.7 | 43.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.7 | 12.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.7 | 2.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.7 | 4.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 4.0 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.7 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.7 | 14.5 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.7 | 3.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.7 | 240.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.6 | 22.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.6 | 4.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.6 | 3.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.6 | 46.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.6 | 1.8 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.6 | 3.0 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.6 | 97.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.6 | 1.7 | GO:0097252 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.6 | 8.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 4.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.5 | 27.4 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.5 | 2.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.5 | 5.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 1.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.5 | 2.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.5 | 2.8 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.5 | 1.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.5 | 4.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 1.8 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.4 | 1.8 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.4 | 0.8 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.4 | 49.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.4 | 2.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.4 | 6.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 2.0 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.4 | 4.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 3.0 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.4 | 4.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.4 | 8.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 11.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.3 | 7.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 29.9 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.3 | 3.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 3.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 4.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.7 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 5.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.3 | 3.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.3 | 2.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.7 | GO:1903538 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.2 | 1.5 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 9.0 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.2 | 2.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.2 | 0.6 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 | 10.0 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 1.7 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 4.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 6.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 9.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.2 | 5.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.6 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 2.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 22.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.2 | 2.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.9 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.2 | 0.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 3.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 20.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 3.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 6.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 9.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 12.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 10.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 2.9 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 3.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 1.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 7.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 11.7 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.1 | 1.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 3.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 9.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.4 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 3.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 5.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 4.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.5 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.1 | 1.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.7 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.8 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 1.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.6 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.1 | 4.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.4 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 1.1 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 1.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 5.3 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 1.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.9 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 2.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.8 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 1.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.8 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.9 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.6 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 32.7 | 196.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 28.5 | 85.6 | GO:0001534 | radial spoke(GO:0001534) |
| 25.0 | 250.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 14.5 | 101.2 | GO:0002177 | manchette(GO:0002177) |
| 10.8 | 21.6 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 10.5 | 241.8 | GO:0036038 | MKS complex(GO:0036038) |
| 7.7 | 76.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 7.3 | 21.8 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 7.1 | 98.9 | GO:0034464 | BBSome(GO:0034464) |
| 5.7 | 63.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 5.4 | 108.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 5.4 | 97.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 5.0 | 85.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 4.8 | 9.7 | GO:0016939 | kinesin II complex(GO:0016939) |
| 4.8 | 147.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 4.1 | 379.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 3.5 | 276.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 3.4 | 64.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 3.3 | 321.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 2.0 | 17.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.4 | 78.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.3 | 63.7 | GO:0031514 | motile cilium(GO:0031514) |
| 1.1 | 7.8 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.9 | 67.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.9 | 6.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 7.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 4.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 21.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.8 | 23.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.8 | 9.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.7 | 4.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 7.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.6 | 191.4 | GO:0005929 | cilium(GO:0005929) |
| 0.6 | 11.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 32.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 4.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 2.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 1.6 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.4 | 3.0 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.4 | 3.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 1.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 4.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.3 | 0.9 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 135.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 9.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 0.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.8 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 2.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 6.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 28.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 9.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 13.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 8.5 | GO:0032153 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 1.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 12.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 17.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 5.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 79.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 5.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 18.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.0 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 3.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 43.8 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 4.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 121.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 11.3 | 158.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 9.5 | 47.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 7.7 | 23.2 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 6.7 | 66.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 6.0 | 36.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 5.3 | 163.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 5.0 | 15.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 4.9 | 29.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 4.7 | 108.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 4.1 | 56.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 4.0 | 112.9 | GO:0031005 | filamin binding(GO:0031005) |
| 3.3 | 16.4 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 2.9 | 90.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 2.9 | 8.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 2.7 | 8.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 2.4 | 106.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 2.3 | 7.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 1.9 | 13.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.8 | 7.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.7 | 184.4 | GO:0043621 | protein self-association(GO:0043621) |
| 1.6 | 71.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.5 | 57.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.4 | 10.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.4 | 8.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 1.4 | 44.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.3 | 2.7 | GO:0015925 | galactosidase activity(GO:0015925) |
| 1.3 | 3.9 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 1.3 | 6.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.2 | 7.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.2 | 5.9 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 1.1 | 21.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 1.1 | 4.5 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.1 | 15.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 1.1 | 9.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.0 | 17.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.9 | 2.8 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.8 | 4.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.7 | 4.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.7 | 31.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 7.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 3.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 34.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.6 | 3.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.6 | 2.4 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.6 | 22.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 39.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.6 | 2.9 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.6 | 2.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 2.8 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.6 | 210.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.6 | 1.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 3.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 24.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.5 | 14.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 18.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.5 | 2.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.5 | 8.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 3.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.5 | 26.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 9.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.4 | 222.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.4 | 1.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 91.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.4 | 2.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.4 | 3.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 7.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 9.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 1.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 1.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 1.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 9.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 7.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 15.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 6.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 2.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 14.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 2.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 5.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 4.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 9.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 3.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.9 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.2 | 4.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 4.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 7.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 36.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 2.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 3.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 8.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 3.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 2.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 11.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 6.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 14.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 2.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 14.0 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 12.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 2.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.0 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 82.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0004583 | alpha-1,3-mannosyltransferase activity(GO:0000033) dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 4.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 22.4 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 5.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.6 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 3.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 50.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.7 | 9.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 42.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.4 | 30.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 6.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 29.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.4 | 4.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 29.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.3 | 11.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 62.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 5.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 10.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 6.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 4.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 7.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 69.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.0 | 27.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.8 | 30.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.5 | 32.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.5 | 10.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 11.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.5 | 14.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.4 | 12.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.4 | 13.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 9.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 14.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 9.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 6.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 4.0 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 3.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 3.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 4.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 4.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 3.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 2.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 7.0 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 6.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 2.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |