Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
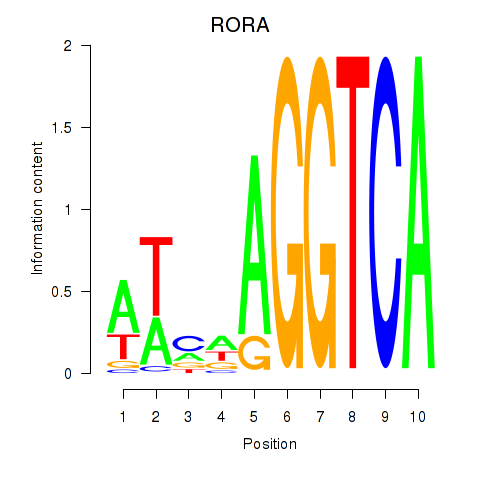
Results for RORA
Z-value: 0.35
Transcription factors associated with RORA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORA
|
ENSG00000069667.16 | RORA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORA | hg38_v1_chr15_-_60592507_60592545 | 0.21 | 2.7e-01 | Click! |
Activity profile of RORA motif
Sorted Z-values of RORA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RORA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_727117 | 0.49 |
ENST00000562141.5
|
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr11_+_73950985 | 0.47 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr1_+_103617427 | 0.43 |
ENST00000423678.2
ENST00000414303.7 |
AMY2A
|
amylase alpha 2A |
| chr11_-_123885627 | 0.42 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr7_+_123601836 | 0.40 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601815 | 0.40 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_+_103750406 | 0.39 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C |
| chr1_+_212565334 | 0.35 |
ENST00000366981.8
ENST00000366987.6 |
ATF3
|
activating transcription factor 3 |
| chr7_-_105679089 | 0.34 |
ENST00000477775.5
|
ATXN7L1
|
ataxin 7 like 1 |
| chr5_+_38258373 | 0.32 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr5_+_38258551 | 0.29 |
ENST00000322350.10
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains |
| chr15_+_43517590 | 0.29 |
ENST00000300231.6
|
MAP1A
|
microtubule associated protein 1A |
| chr16_+_727246 | 0.28 |
ENST00000561546.5
ENST00000564545.1 ENST00000567414.5 ENST00000568141.5 |
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr1_+_46175079 | 0.27 |
ENST00000372003.6
|
TSPAN1
|
tetraspanin 1 |
| chr3_-_24494842 | 0.27 |
ENST00000646209.2
|
THRB
|
thyroid hormone receptor beta |
| chr19_-_48637338 | 0.26 |
ENST00000601104.1
ENST00000222122.10 |
DBP
|
D-box binding PAR bZIP transcription factor |
| chr7_+_100969565 | 0.25 |
ENST00000536621.6
ENST00000379442.7 |
MUC12
|
mucin 12, cell surface associated |
| chr3_-_123620571 | 0.25 |
ENST00000583087.5
|
MYLK
|
myosin light chain kinase |
| chr14_-_58427134 | 0.25 |
ENST00000555930.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr3_-_123620496 | 0.25 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_24494791 | 0.23 |
ENST00000431815.5
ENST00000356447.9 ENST00000418247.1 ENST00000416420.5 ENST00000396671.7 |
THRB
|
thyroid hormone receptor beta |
| chr6_-_106629472 | 0.23 |
ENST00000369063.8
|
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr16_+_28292485 | 0.21 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr11_-_7963646 | 0.20 |
ENST00000328600.3
|
NLRP10
|
NLR family pyrin domain containing 10 |
| chr14_-_70188967 | 0.20 |
ENST00000381269.6
ENST00000357887.7 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr14_-_70188937 | 0.19 |
ENST00000356921.7
|
SLC8A3
|
solute carrier family 8 member A3 |
| chr12_-_6606642 | 0.19 |
ENST00000545584.2
ENST00000545942.6 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr12_-_39443390 | 0.19 |
ENST00000361961.7
|
KIF21A
|
kinesin family member 21A |
| chr10_+_102419189 | 0.17 |
ENST00000432590.5
|
FBXL15
|
F-box and leucine rich repeat protein 15 |
| chr8_-_133060347 | 0.17 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr16_+_726936 | 0.17 |
ENST00000549114.5
ENST00000341413.8 ENST00000562187.1 ENST00000564537.5 ENST00000389703.8 |
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr17_-_2025289 | 0.17 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chr15_+_76059973 | 0.17 |
ENST00000388942.8
|
TMEM266
|
transmembrane protein 266 |
| chr9_-_39239174 | 0.16 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr21_-_42395943 | 0.16 |
ENST00000398405.5
|
TMPRSS3
|
transmembrane serine protease 3 |
| chr10_-_63269057 | 0.16 |
ENST00000542921.5
|
JMJD1C
|
jumonji domain containing 1C |
| chr10_-_92291063 | 0.16 |
ENST00000265997.5
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_-_24494875 | 0.16 |
ENST00000644321.1
|
THRB
|
thyroid hormone receptor beta |
| chr19_+_15949008 | 0.15 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr19_-_49808639 | 0.15 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr18_-_58629084 | 0.15 |
ENST00000361673.4
|
ALPK2
|
alpha kinase 2 |
| chr19_+_1248553 | 0.14 |
ENST00000586757.5
ENST00000300952.6 ENST00000682408.1 |
MIDN
|
midnolin |
| chr2_-_206159410 | 0.14 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr11_-_64759967 | 0.14 |
ENST00000377432.7
ENST00000164139.4 |
PYGM
|
glycogen phosphorylase, muscle associated |
| chr2_+_108621260 | 0.14 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr1_-_235649734 | 0.14 |
ENST00000391854.7
|
GNG4
|
G protein subunit gamma 4 |
| chr12_-_16608073 | 0.14 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr21_-_42396043 | 0.12 |
ENST00000433957.7
ENST00000644384.2 ENST00000652415.1 ENST00000398397.3 |
TMPRSS3
|
transmembrane serine protease 3 |
| chr1_-_157138474 | 0.11 |
ENST00000326786.4
|
ETV3
|
ETS variant transcription factor 3 |
| chr19_-_51065067 | 0.11 |
ENST00000595547.5
ENST00000335422.3 ENST00000595793.6 ENST00000596955.1 |
KLK13
|
kallikrein related peptidase 13 |
| chr19_-_55624563 | 0.10 |
ENST00000325351.5
ENST00000591479.1 |
ZNF784
|
zinc finger protein 784 |
| chr5_-_137736066 | 0.10 |
ENST00000309755.9
|
KLHL3
|
kelch like family member 3 |
| chr12_+_93569814 | 0.10 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr14_-_23286082 | 0.10 |
ENST00000357460.7
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr6_+_52186373 | 0.10 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chr12_+_109139397 | 0.09 |
ENST00000377854.9
ENST00000377848.7 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr6_-_96897853 | 0.09 |
ENST00000316149.8
|
NDUFAF4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr5_-_132011811 | 0.09 |
ENST00000379255.5
ENST00000430403.5 ENST00000357096.5 |
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr5_+_140647802 | 0.09 |
ENST00000417647.7
ENST00000507593.5 ENST00000508301.5 |
IK
|
IK cytokine |
| chr14_+_103121457 | 0.09 |
ENST00000333007.8
|
TNFAIP2
|
TNF alpha induced protein 2 |
| chr15_-_51971740 | 0.09 |
ENST00000315141.5
ENST00000299601.10 |
LEO1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr16_+_31355165 | 0.09 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr19_-_3546306 | 0.09 |
ENST00000398558.8
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr11_+_7513966 | 0.09 |
ENST00000299492.9
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr1_-_154192058 | 0.09 |
ENST00000271850.11
ENST00000368530.7 ENST00000651641.1 |
TPM3
|
tropomyosin 3 |
| chr18_-_12377200 | 0.09 |
ENST00000269143.8
|
AFG3L2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr16_+_82056423 | 0.08 |
ENST00000568090.5
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr15_-_51243011 | 0.08 |
ENST00000405913.7
ENST00000559878.5 |
CYP19A1
|
cytochrome P450 family 19 subfamily A member 1 |
| chr2_+_233307806 | 0.08 |
ENST00000447536.5
ENST00000409110.6 |
SAG
|
S-antigen visual arrestin |
| chr15_-_74725370 | 0.08 |
ENST00000567032.5
ENST00000564596.5 ENST00000566503.1 ENST00000395049.8 ENST00000379727.8 ENST00000617691.4 ENST00000395048.6 |
CYP1A1
|
cytochrome P450 family 1 subfamily A member 1 |
| chr1_-_91021996 | 0.08 |
ENST00000337393.10
|
ZNF644
|
zinc finger protein 644 |
| chr10_-_101818405 | 0.08 |
ENST00000357797.9
ENST00000370094.7 |
OGA
|
O-GlcNAcase |
| chr3_-_24495532 | 0.08 |
ENST00000643772.1
ENST00000642307.1 ENST00000645139.1 |
THRB
|
thyroid hormone receptor beta |
| chr5_+_170861990 | 0.08 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr9_+_131228109 | 0.08 |
ENST00000498010.2
ENST00000476004.5 ENST00000528406.1 |
NUP214
|
nucleoporin 214 |
| chrX_-_130165699 | 0.08 |
ENST00000676328.1
ENST00000675857.1 ENST00000675427.1 ENST00000675092.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr12_+_93570381 | 0.08 |
ENST00000549206.5
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr19_-_54313074 | 0.07 |
ENST00000486742.2
ENST00000432233.8 |
LILRA5
|
leukocyte immunoglobulin like receptor A5 |
| chrX_+_10156960 | 0.07 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr1_+_20070156 | 0.07 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V |
| chr6_-_88963409 | 0.07 |
ENST00000369475.7
ENST00000538899.2 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chrX_-_150898592 | 0.07 |
ENST00000355149.8
ENST00000466436.5 ENST00000370377.8 |
CD99L2
|
CD99 molecule like 2 |
| chr2_-_148020689 | 0.07 |
ENST00000457954.5
ENST00000392857.10 ENST00000540442.5 ENST00000535373.5 |
ORC4
|
origin recognition complex subunit 4 |
| chr5_+_153490655 | 0.07 |
ENST00000518142.5
ENST00000285900.10 |
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr4_+_105146868 | 0.07 |
ENST00000380013.9
ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr17_+_62370218 | 0.07 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr3_+_130431463 | 0.06 |
ENST00000512836.5
|
COL6A5
|
collagen type VI alpha 5 chain |
| chrX_-_109482075 | 0.06 |
ENST00000218006.3
|
GUCY2F
|
guanylate cyclase 2F, retinal |
| chr10_-_92243246 | 0.06 |
ENST00000412050.8
ENST00000614585.4 |
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_74206384 | 0.06 |
ENST00000678623.1
ENST00000678731.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr1_-_84574407 | 0.06 |
ENST00000370630.6
|
CTBS
|
chitobiase |
| chr7_-_151080833 | 0.06 |
ENST00000353841.6
ENST00000482571.2 |
FASTK
|
Fas activated serine/threonine kinase |
| chr18_-_21704763 | 0.06 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr4_-_21697755 | 0.06 |
ENST00000382148.7
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr1_+_45913647 | 0.06 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr11_-_112086727 | 0.06 |
ENST00000504148.3
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr16_+_2026834 | 0.06 |
ENST00000424542.7
ENST00000432365.6 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr5_-_132011794 | 0.05 |
ENST00000650697.1
|
ACSL6
|
acyl-CoA synthetase long chain family member 6 |
| chr1_+_28887166 | 0.05 |
ENST00000347529.7
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr16_-_1884231 | 0.05 |
ENST00000563416.3
ENST00000633813.1 ENST00000470044.5 |
LINC00254
MEIOB
|
long intergenic non-protein coding RNA 254 meiosis specific with OB-fold |
| chrX_-_150898779 | 0.05 |
ENST00000613030.4
ENST00000437787.6 |
CD99L2
|
CD99 molecule like 2 |
| chr3_+_179604785 | 0.05 |
ENST00000611971.4
ENST00000493866.5 ENST00000259037.8 ENST00000472629.5 ENST00000482604.5 |
NDUFB5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr6_-_88963573 | 0.05 |
ENST00000369485.9
|
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr1_+_51236252 | 0.05 |
ENST00000242719.4
|
RNF11
|
ring finger protein 11 |
| chr3_-_42804451 | 0.05 |
ENST00000418900.6
ENST00000321331.12 ENST00000430190.5 ENST00000648550.1 |
HIGD1A
ENSG00000280571.2
|
HIG1 hypoxia inducible domain family member 1A novel protein |
| chr17_-_2711633 | 0.05 |
ENST00000435359.5
|
CLUH
|
clustered mitochondria homolog |
| chr5_+_73813518 | 0.05 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chr7_-_45111673 | 0.05 |
ENST00000461363.1
ENST00000258770.8 ENST00000495078.1 ENST00000494076.5 ENST00000478532.5 ENST00000361278.7 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr9_-_128204218 | 0.04 |
ENST00000634901.1
ENST00000372948.7 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr5_-_88785493 | 0.04 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chrX_-_130165664 | 0.04 |
ENST00000535724.6
ENST00000346424.6 ENST00000676436.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chrX_-_30309387 | 0.04 |
ENST00000378970.5
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr11_+_65111845 | 0.04 |
ENST00000526809.5
ENST00000524986.5 ENST00000534371.5 ENST00000279263.14 ENST00000525385.5 ENST00000345348.9 ENST00000531321.5 ENST00000529414.5 ENST00000526085.5 ENST00000530750.5 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr3_-_50322733 | 0.04 |
ENST00000428028.1
ENST00000357750.9 |
HYAL2
|
hyaluronidase 2 |
| chr3_+_141386393 | 0.04 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_-_6440980 | 0.04 |
ENST00000265983.8
ENST00000615166.1 |
HPX
|
hemopexin |
| chr7_-_151080787 | 0.04 |
ENST00000540185.5
ENST00000297532.11 |
FASTK
|
Fas activated serine/threonine kinase |
| chr1_-_153550083 | 0.04 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr2_+_37925256 | 0.04 |
ENST00000354545.8
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr16_-_49664225 | 0.04 |
ENST00000535559.5
|
ZNF423
|
zinc finger protein 423 |
| chr5_-_140647590 | 0.04 |
ENST00000252102.9
ENST00000512088.1 |
NDUFA2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr22_-_23767876 | 0.04 |
ENST00000520222.1
ENST00000401675.7 ENST00000484558.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr3_-_50322759 | 0.03 |
ENST00000442581.1
ENST00000447092.5 |
HYAL2
|
hyaluronidase 2 |
| chr2_+_197500371 | 0.03 |
ENST00000409468.1
ENST00000233893.10 |
HSPE1
|
heat shock protein family E (Hsp10) member 1 |
| chr14_-_103522696 | 0.03 |
ENST00000553878.5
ENST00000348956.7 ENST00000557530.1 |
CKB
|
creatine kinase B |
| chr5_-_127073467 | 0.03 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr20_+_18145083 | 0.03 |
ENST00000489634.2
|
KAT14
|
lysine acetyltransferase 14 |
| chr2_+_197500398 | 0.03 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr6_+_106629594 | 0.03 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA amidotransferase subunit QRSL1 |
| chr3_+_155870623 | 0.03 |
ENST00000295920.7
ENST00000496455.7 |
GMPS
|
guanine monophosphate synthase |
| chr7_-_101180215 | 0.03 |
ENST00000455377.5
ENST00000443096.1 ENST00000300303.7 |
NAT16
|
N-acetyltransferase 16 (putative) |
| chr6_+_106629560 | 0.03 |
ENST00000369046.8
|
QRSL1
|
glutaminyl-tRNA amidotransferase subunit QRSL1 |
| chr20_+_34977625 | 0.03 |
ENST00000618182.6
|
MYH7B
|
myosin heavy chain 7B |
| chr1_+_36224410 | 0.03 |
ENST00000469141.6
ENST00000648638.1 ENST00000354618.10 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr8_+_119208322 | 0.03 |
ENST00000614891.5
|
MAL2
|
mal, T cell differentiation protein 2 |
| chrX_-_130165825 | 0.03 |
ENST00000675240.1
ENST00000319908.8 ENST00000674546.1 ENST00000287295.8 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr10_-_126670686 | 0.03 |
ENST00000488181.3
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr11_-_67504252 | 0.03 |
ENST00000533391.5
ENST00000534749.5 ENST00000532703.5 |
PITPNM1
|
phosphatidylinositol transfer protein membrane associated 1 |
| chr5_-_137735997 | 0.02 |
ENST00000505853.1
|
KLHL3
|
kelch like family member 3 |
| chr11_+_83156988 | 0.02 |
ENST00000298281.8
ENST00000530660.5 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr19_+_35030438 | 0.02 |
ENST00000415950.5
ENST00000262631.11 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr1_-_31938302 | 0.02 |
ENST00000647444.2
|
PTP4A2
|
protein tyrosine phosphatase 4A2 |
| chr2_+_10420021 | 0.02 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin like 1 |
| chr1_+_160177386 | 0.02 |
ENST00000470705.1
|
ATP1A4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr2_-_206159509 | 0.02 |
ENST00000423725.5
|
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr17_-_4555371 | 0.02 |
ENST00000254718.9
ENST00000381556.6 |
MYBBP1A
|
MYB binding protein 1a |
| chr10_+_70815889 | 0.02 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr11_-_33717409 | 0.02 |
ENST00000651485.1
|
CD59
|
CD59 molecule (CD59 blood group) |
| chr3_+_113747022 | 0.02 |
ENST00000273398.8
ENST00000496747.5 ENST00000475322.1 |
ATP6V1A
|
ATPase H+ transporting V1 subunit A |
| chr5_-_141958174 | 0.02 |
ENST00000231484.4
|
PCDH12
|
protocadherin 12 |
| chr2_-_197499857 | 0.02 |
ENST00000428204.6
ENST00000678170.1 ENST00000676933.1 ENST00000678621.1 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr17_-_46192767 | 0.02 |
ENST00000262419.10
|
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr11_+_118401586 | 0.01 |
ENST00000649464.1
ENST00000648261.1 ENST00000300688.8 ENST00000533172.5 ENST00000524422.1 |
ENSG00000285827.1
ATP5MG
|
novel protein ATP synthase membrane subunit g |
| chr7_-_128343823 | 0.01 |
ENST00000415472.6
ENST00000478061.5 ENST00000223073.6 ENST00000459726.1 |
RBM28
|
RNA binding motif protein 28 |
| chr1_-_185317234 | 0.01 |
ENST00000367498.8
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr20_+_38962299 | 0.01 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr5_+_96936071 | 0.01 |
ENST00000231368.10
|
LNPEP
|
leucyl and cystinyl aminopeptidase |
| chr2_+_161160299 | 0.01 |
ENST00000440506.5
ENST00000429217.5 ENST00000406287.5 ENST00000402568.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr2_-_197499826 | 0.01 |
ENST00000439605.2
ENST00000388968.8 ENST00000418022.2 |
HSPD1
|
heat shock protein family D (Hsp60) member 1 |
| chr11_-_129192198 | 0.01 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_18438767 | 0.01 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr17_+_42844573 | 0.01 |
ENST00000253799.8
ENST00000452774.2 |
AOC2
|
amine oxidase copper containing 2 |
| chr8_+_37695774 | 0.01 |
ENST00000331569.6
|
ZNF703
|
zinc finger protein 703 |
| chr17_-_51120855 | 0.01 |
ENST00000618113.4
ENST00000357122.8 ENST00000262013.12 |
SPAG9
|
sperm associated antigen 9 |
| chr11_-_129192291 | 0.01 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_179604628 | 0.01 |
ENST00000476781.6
ENST00000259038.6 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr18_+_9102671 | 0.01 |
ENST00000497577.2
ENST00000318388.11 ENST00000579126.5 ENST00000578850.1 |
NDUFV2
ENSG00000265257.5
|
NADH:ubiquinone oxidoreductase core subunit V2 novel transcript |
| chr11_-_132943092 | 0.00 |
ENST00000612177.4
ENST00000541867.5 |
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr19_-_39435627 | 0.00 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr1_+_161202147 | 0.00 |
ENST00000392179.5
ENST00000678511.1 ENST00000677453.1 ENST00000678783.1 ENST00000679218.1 ENST00000676972.1 |
NDUFS2
|
NADH:ubiquinone oxidoreductase core subunit S2 |
| chr20_+_54475647 | 0.00 |
ENST00000395939.5
|
DOK5
|
docking protein 5 |
| chr3_-_179604648 | 0.00 |
ENST00000392659.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr1_-_205750167 | 0.00 |
ENST00000367142.5
|
NUCKS1
|
nuclear casein kinase and cyclin dependent kinase substrate 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.7 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |