|
chr19_-_50968125
Show fit
|
10.03 |
ENST00000594641.1
|
KLK6
|
kallikrein related peptidase 6
|
|
chr19_-_50953063
Show fit
|
6.62 |
ENST00000391809.6
|
KLK5
|
kallikrein related peptidase 5
|
|
chr19_-_50953093
Show fit
|
6.59 |
ENST00000593428.5
|
KLK5
|
kallikrein related peptidase 5
|
|
chr19_-_50952942
Show fit
|
6.57 |
ENST00000594846.1
ENST00000336334.8
|
KLK5
|
kallikrein related peptidase 5
|
|
chr6_+_151325665
Show fit
|
5.85 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12
|
|
chr18_-_23662810
Show fit
|
4.57 |
ENST00000322980.13
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr4_+_74365136
Show fit
|
4.19 |
ENST00000244869.3
|
EREG
|
epiregulin
|
|
chr4_+_8229170
Show fit
|
3.54 |
ENST00000511002.6
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr18_-_23662868
Show fit
|
3.51 |
ENST00000586087.1
ENST00000592179.6
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr2_+_233195433
Show fit
|
3.41 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase D
|
|
chr10_+_17229267
Show fit
|
3.18 |
ENST00000224237.9
|
VIM
|
vimentin
|
|
chr19_-_18941117
Show fit
|
3.00 |
ENST00000600077.5
|
HOMER3
|
homer scaffold protein 3
|
|
chr12_-_121793668
Show fit
|
2.86 |
ENST00000267205.7
|
RHOF
|
ras homolog family member F, filopodia associated
|
|
chr17_+_64449037
Show fit
|
2.72 |
ENST00000615220.4
ENST00000619286.5
ENST00000612535.4
ENST00000616498.4
|
MILR1
|
mast cell immunoglobulin like receptor 1
|
|
chr1_-_16980607
Show fit
|
2.69 |
ENST00000375535.4
|
MFAP2
|
microfibril associated protein 2
|
|
chr9_+_128411715
Show fit
|
2.51 |
ENST00000420034.5
ENST00000372842.5
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr19_-_18941184
Show fit
|
2.51 |
ENST00000594794.5
ENST00000392351.8
ENST00000596482.5
|
HOMER3
|
homer scaffold protein 3
|
|
chr8_+_32548303
Show fit
|
2.44 |
ENST00000650967.1
|
NRG1
|
neuregulin 1
|
|
chr16_+_397209
Show fit
|
2.40 |
ENST00000382940.8
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
|
chr16_-_4538819
Show fit
|
2.31 |
ENST00000564828.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr8_+_32548210
Show fit
|
2.29 |
ENST00000523079.5
ENST00000650919.1
|
NRG1
|
neuregulin 1
|
|
chrX_+_17737443
Show fit
|
2.21 |
ENST00000398080.5
ENST00000380045.7
ENST00000380043.7
ENST00000380041.8
|
SCML1
|
Scm polycomb group protein like 1
|
|
chr16_+_58500047
Show fit
|
2.19 |
ENST00000566192.5
ENST00000565088.5
ENST00000568640.5
|
NDRG4
|
NDRG family member 4
|
|
chr5_+_151212117
Show fit
|
2.17 |
ENST00000523466.5
|
GM2A
|
GM2 ganglioside activator
|
|
chr16_-_4538761
Show fit
|
2.14 |
ENST00000567695.6
ENST00000562334.5
ENST00000562579.5
ENST00000563507.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr16_-_4538469
Show fit
|
2.09 |
ENST00000588381.1
ENST00000563332.6
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr16_+_397226
Show fit
|
2.01 |
ENST00000433358.5
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
|
chr16_+_58500135
Show fit
|
2.00 |
ENST00000563978.5
ENST00000569923.5
ENST00000356752.8
ENST00000563799.5
ENST00000562999.5
ENST00000570248.5
ENST00000562731.5
ENST00000568424.1
|
NDRG4
|
NDRG family member 4
|
|
chr18_+_36297661
Show fit
|
1.88 |
ENST00000257209.8
ENST00000590592.5
ENST00000359247.8
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr10_-_99235783
Show fit
|
1.87 |
ENST00000370546.5
ENST00000614306.1
|
HPSE2
|
heparanase 2 (inactive)
|
|
chr7_-_24757413
Show fit
|
1.80 |
ENST00000645220.1
ENST00000409970.6
|
GSDME
|
gasdermin E
|
|
chr10_-_3785225
Show fit
|
1.76 |
ENST00000542957.1
|
KLF6
|
Kruppel like factor 6
|
|
chr12_-_52680398
Show fit
|
1.75 |
ENST00000252244.3
|
KRT1
|
keratin 1
|
|
chr10_-_3785197
Show fit
|
1.70 |
ENST00000497571.6
|
KLF6
|
Kruppel like factor 6
|
|
chr7_-_128405930
Show fit
|
1.67 |
ENST00000470772.5
ENST00000480861.5
ENST00000496200.5
|
IMPDH1
|
inosine monophosphate dehydrogenase 1
|
|
chr2_-_85410336
Show fit
|
1.67 |
ENST00000263867.9
ENST00000409921.5
|
CAPG
|
capping actin protein, gelsolin like
|
|
chr19_-_50719761
Show fit
|
1.60 |
ENST00000293441.6
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr1_+_65525641
Show fit
|
1.53 |
ENST00000344610.12
ENST00000616738.4
|
LEPR
|
leptin receptor
|
|
chr19_-_38735405
Show fit
|
1.53 |
ENST00000597987.5
ENST00000595177.1
|
CAPN12
|
calpain 12
|
|
chr16_+_397183
Show fit
|
1.51 |
ENST00000620944.4
ENST00000621774.4
ENST00000219479.7
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
|
chr9_-_104928139
Show fit
|
1.51 |
ENST00000423487.6
ENST00000374733.1
ENST00000374736.8
ENST00000678995.1
|
ABCA1
|
ATP binding cassette subfamily A member 1
|
|
chr17_-_41060109
Show fit
|
1.51 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3
|
|
chrX_+_135032346
Show fit
|
1.48 |
ENST00000257013.9
|
RTL8C
|
retrotransposon Gag like 8C
|
|
chr21_+_36135071
Show fit
|
1.47 |
ENST00000290354.6
|
CBR3
|
carbonyl reductase 3
|
|
chr17_-_41481140
Show fit
|
1.44 |
ENST00000246639.6
ENST00000393989.1
|
KRT35
|
keratin 35
|
|
chr2_+_28395511
Show fit
|
1.42 |
ENST00000436647.1
|
FOSL2
|
FOS like 2, AP-1 transcription factor subunit
|
|
chr18_+_74499939
Show fit
|
1.42 |
ENST00000584768.5
|
CNDP2
|
carnosine dipeptidase 2
|
|
chr12_-_119803383
Show fit
|
1.40 |
ENST00000392520.2
ENST00000678677.1
ENST00000679249.1
ENST00000676849.1
|
CIT
|
citron rho-interacting serine/threonine kinase
|
|
chr1_-_228416841
Show fit
|
1.40 |
ENST00000366698.7
ENST00000457345.2
|
TRIM17
|
tripartite motif containing 17
|
|
chr12_-_7936177
Show fit
|
1.37 |
ENST00000544291.1
ENST00000075120.12
|
SLC2A3
|
solute carrier family 2 member 3
|
|
chr9_+_35906179
Show fit
|
1.37 |
ENST00000354323.3
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr3_-_42702778
Show fit
|
1.32 |
ENST00000457462.5
ENST00000441594.6
|
HHATL
|
hedgehog acyltransferase like
|
|
chr20_-_38165261
Show fit
|
1.32 |
ENST00000361475.7
|
TGM2
|
transglutaminase 2
|
|
chr3_-_169769542
Show fit
|
1.26 |
ENST00000330368.3
|
ACTRT3
|
actin related protein T3
|
|
chr10_-_33335074
Show fit
|
1.23 |
ENST00000432372.6
|
NRP1
|
neuropilin 1
|
|
chr1_-_228416626
Show fit
|
1.22 |
ENST00000355586.4
ENST00000520264.1
ENST00000479800.1
ENST00000295033.7
|
TRIM17
|
tripartite motif containing 17
|
|
chr8_+_32548267
Show fit
|
1.21 |
ENST00000356819.7
|
NRG1
|
neuregulin 1
|
|
chr14_+_104689588
Show fit
|
1.19 |
ENST00000330634.11
ENST00000392634.9
ENST00000675482.1
ENST00000398337.8
|
INF2
|
inverted formin 2
|
|
chr12_-_94650506
Show fit
|
1.17 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr19_-_14112641
Show fit
|
1.15 |
ENST00000536649.5
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha
|
|
chr1_-_161223559
Show fit
|
1.12 |
ENST00000469730.2
ENST00000463273.5
ENST00000464492.5
ENST00000367990.7
ENST00000470459.6
ENST00000463812.1
ENST00000468465.5
|
APOA2
|
apolipoprotein A2
|
|
chr20_+_43667019
Show fit
|
1.11 |
ENST00000396863.8
|
MYBL2
|
MYB proto-oncogene like 2
|
|
chr15_+_63189554
Show fit
|
1.10 |
ENST00000559006.1
ENST00000321437.9
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr3_+_155080088
Show fit
|
1.10 |
ENST00000462745.5
|
MME
|
membrane metalloendopeptidase
|
|
chr17_-_75844334
Show fit
|
1.08 |
ENST00000592386.5
ENST00000412096.6
ENST00000586147.1
ENST00000207549.9
|
UNC13D
|
unc-13 homolog D
|
|
chr10_-_33334898
Show fit
|
1.06 |
ENST00000395995.5
|
NRP1
|
neuropilin 1
|
|
chr10_-_3785179
Show fit
|
1.05 |
ENST00000469435.1
|
KLF6
|
Kruppel like factor 6
|
|
chr12_+_26195647
Show fit
|
1.05 |
ENST00000535504.1
|
SSPN
|
sarcospan
|
|
chr10_-_33334625
Show fit
|
1.04 |
ENST00000374875.5
ENST00000374822.8
ENST00000374867.7
|
NRP1
|
neuropilin 1
|
|
chr20_+_43667105
Show fit
|
1.03 |
ENST00000217026.5
|
MYBL2
|
MYB proto-oncogene like 2
|
|
chr3_+_155079847
Show fit
|
1.00 |
ENST00000615825.2
|
MME
|
membrane metalloendopeptidase
|
|
chr8_-_143541425
Show fit
|
0.99 |
ENST00000262577.6
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr22_-_35961623
Show fit
|
0.98 |
ENST00000408983.2
|
RBFOX2
|
RNA binding fox-1 homolog 2
|
|
chr11_+_847197
Show fit
|
0.94 |
ENST00000532375.5
ENST00000346501.8
|
TSPAN4
|
tetraspanin 4
|
|
chr17_-_41065879
Show fit
|
0.93 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chrX_-_132218124
Show fit
|
0.93 |
ENST00000342983.6
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr1_+_101238090
Show fit
|
0.92 |
ENST00000475289.2
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_+_60422483
Show fit
|
0.92 |
ENST00000269127.5
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr1_+_109548567
Show fit
|
0.91 |
ENST00000369851.7
|
GNAI3
|
G protein subunit alpha i3
|
|
chrX_+_154585120
Show fit
|
0.89 |
ENST00000593606.3
ENST00000599837.3
|
CTAG1A
|
cancer/testis antigen 1A
|
|
chr21_-_46228751
Show fit
|
0.89 |
ENST00000450351.1
ENST00000397728.8
ENST00000522411.5
ENST00000356396.8
ENST00000457828.6
|
LSS
|
lanosterol synthase
|
|
chr16_+_28878480
Show fit
|
0.87 |
ENST00000395503.9
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1
|
|
chr11_-_59668981
Show fit
|
0.86 |
ENST00000300146.10
|
PATL1
|
PAT1 homolog 1, processing body mRNA decay factor
|
|
chr10_-_119596954
Show fit
|
0.86 |
ENST00000436547.7
|
TIAL1
|
TIA1 cytotoxic granule associated RNA binding protein like 1
|
|
chr5_-_151924824
Show fit
|
0.85 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1
|
|
chr17_+_2055094
Show fit
|
0.84 |
ENST00000399849.4
ENST00000619757.5
|
HIC1
|
HIC ZBTB transcriptional repressor 1
|
|
chr3_-_195909711
Show fit
|
0.83 |
ENST00000333602.14
|
TNK2
|
tyrosine kinase non receptor 2
|
|
chr3_+_155079663
Show fit
|
0.83 |
ENST00000460393.6
|
MME
|
membrane metalloendopeptidase
|
|
chr10_-_99235846
Show fit
|
0.83 |
ENST00000370552.8
ENST00000370549.5
ENST00000628193.2
|
HPSE2
|
heparanase 2 (inactive)
|
|
chrX_-_154619282
Show fit
|
0.82 |
ENST00000328435.3
ENST00000359887.4
|
CTAG1B
|
cancer/testis antigen 1B
|
|
chr8_+_26291758
Show fit
|
0.82 |
ENST00000522535.5
ENST00000665949.1
|
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha
|
|
chr17_+_40318237
Show fit
|
0.82 |
ENST00000394089.6
ENST00000425707.7
|
RARA
|
retinoic acid receptor alpha
|
|
chr8_+_32548590
Show fit
|
0.81 |
ENST00000652588.1
ENST00000521670.5
ENST00000287842.7
|
NRG1
|
neuregulin 1
|
|
chr3_+_155079911
Show fit
|
0.81 |
ENST00000675418.2
|
MME
|
membrane metalloendopeptidase
|
|
chr15_-_89221558
Show fit
|
0.80 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1
|
|
chr18_-_46559289
Show fit
|
0.80 |
ENST00000419859.1
ENST00000582408.6
ENST00000300591.11
|
LOXHD1
|
lipoxygenase homology domains 1
|
|
chr5_-_142686079
Show fit
|
0.79 |
ENST00000337706.7
|
FGF1
|
fibroblast growth factor 1
|
|
chr2_+_227472363
Show fit
|
0.78 |
ENST00000409315.5
ENST00000373671.7
ENST00000409171.5
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr16_+_27313879
Show fit
|
0.78 |
ENST00000562142.5
ENST00000561742.5
ENST00000543915.6
ENST00000395762.7
ENST00000563002.5
|
IL4R
|
interleukin 4 receptor
|
|
chr17_+_60421697
Show fit
|
0.78 |
ENST00000474834.5
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr12_-_120250145
Show fit
|
0.78 |
ENST00000458477.6
|
PXN
|
paxillin
|
|
chr16_+_28878382
Show fit
|
0.75 |
ENST00000357084.7
|
ATP2A1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1
|
|
chr2_-_32011001
Show fit
|
0.74 |
ENST00000404530.6
|
MEMO1
|
mediator of cell motility 1
|
|
chr4_+_146638890
Show fit
|
0.73 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr7_-_712437
Show fit
|
0.72 |
ENST00000360274.8
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta
|
|
chr3_-_39154558
Show fit
|
0.71 |
ENST00000514182.1
|
CSRNP1
|
cysteine and serine rich nuclear protein 1
|
|
chr12_+_6444932
Show fit
|
0.71 |
ENST00000266557.4
|
CD27
|
CD27 molecule
|
|
chr8_-_59119121
Show fit
|
0.70 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box
|
|
chr12_-_12562344
Show fit
|
0.70 |
ENST00000228862.3
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr20_+_59604527
Show fit
|
0.68 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr1_-_155978144
Show fit
|
0.68 |
ENST00000313695.11
ENST00000497907.5
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr12_+_56021316
Show fit
|
0.68 |
ENST00000547791.2
|
IKZF4
|
IKAROS family zinc finger 4
|
|
chrX_+_47223009
Show fit
|
0.68 |
ENST00000518022.5
ENST00000276052.10
|
CDK16
|
cyclin dependent kinase 16
|
|
chrX_-_126166273
Show fit
|
0.67 |
ENST00000360028.4
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12 like 2
|
|
chr8_+_32548661
Show fit
|
0.66 |
ENST00000650980.1
ENST00000405005.7
|
NRG1
|
neuregulin 1
|
|
chr22_-_37580075
Show fit
|
0.65 |
ENST00000215886.6
|
LGALS2
|
galectin 2
|
|
chr19_-_40714641
Show fit
|
0.63 |
ENST00000678467.1
|
COQ8B
|
coenzyme Q8B
|
|
chr17_-_33293247
Show fit
|
0.63 |
ENST00000225823.7
|
ASIC2
|
acid sensing ion channel subunit 2
|
|
chr20_+_49982969
Show fit
|
0.62 |
ENST00000244050.3
|
SNAI1
|
snail family transcriptional repressor 1
|
|
chr12_+_6828377
Show fit
|
0.62 |
ENST00000290510.10
|
P3H3
|
prolyl 3-hydroxylase 3
|
|
chrX_-_13734575
Show fit
|
0.62 |
ENST00000519885.5
ENST00000458511.7
ENST00000380579.6
ENST00000683983.1
ENST00000683569.1
ENST00000359680.9
|
TRAPPC2
|
trafficking protein particle complex 2
|
|
chr2_-_100417608
Show fit
|
0.62 |
ENST00000264249.8
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr11_+_128693887
Show fit
|
0.61 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor
|
|
chr20_+_833781
Show fit
|
0.58 |
ENST00000381939.1
|
FAM110A
|
family with sequence similarity 110 member A
|
|
chr7_-_3043838
Show fit
|
0.57 |
ENST00000356408.3
ENST00000396946.9
|
CARD11
|
caspase recruitment domain family member 11
|
|
chr18_+_33578213
Show fit
|
0.57 |
ENST00000681521.1
ENST00000269197.12
|
ASXL3
|
ASXL transcriptional regulator 3
|
|
chr12_+_118016690
Show fit
|
0.57 |
ENST00000537315.5
ENST00000454402.7
ENST00000484086.6
ENST00000420967.5
ENST00000392542.6
ENST00000535092.1
|
RFC5
|
replication factor C subunit 5
|
|
chr8_+_38404363
Show fit
|
0.57 |
ENST00000527175.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2
|
|
chr12_+_62260338
Show fit
|
0.57 |
ENST00000353364.7
ENST00000549523.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chrX_-_120115909
Show fit
|
0.56 |
ENST00000217999.3
|
RHOXF1
|
Rhox homeobox family member 1
|
|
chrX_+_103330206
Show fit
|
0.55 |
ENST00000372666.1
ENST00000332431.5
|
TCEAL7
|
transcription elongation factor A like 7
|
|
chr19_+_40467145
Show fit
|
0.55 |
ENST00000338932.7
|
SPTBN4
|
spectrin beta, non-erythrocytic 4
|
|
chr17_+_79778135
Show fit
|
0.54 |
ENST00000310942.9
ENST00000269399.5
|
CBX2
|
chromobox 2
|
|
chr12_+_26195543
Show fit
|
0.54 |
ENST00000242729.7
|
SSPN
|
sarcospan
|
|
chr3_+_141487008
Show fit
|
0.53 |
ENST00000286364.9
ENST00000452898.2
|
RASA2
|
RAS p21 protein activator 2
|
|
chr2_-_53859929
Show fit
|
0.53 |
ENST00000394705.3
ENST00000406625.6
|
GPR75
ASB3
|
G protein-coupled receptor 75
ankyrin repeat and SOCS box containing 3
|
|
chr16_+_21158653
Show fit
|
0.51 |
ENST00000572258.5
ENST00000233047.9
ENST00000261388.7
ENST00000451578.6
ENST00000572599.5
ENST00000577162.1
|
TMEM159
|
transmembrane protein 159
|
|
chr4_-_95548956
Show fit
|
0.51 |
ENST00000513796.5
ENST00000453304.6
|
UNC5C
|
unc-5 netrin receptor C
|
|
chr12_+_26195313
Show fit
|
0.51 |
ENST00000422622.3
|
SSPN
|
sarcospan
|
|
chr3_+_8733779
Show fit
|
0.50 |
ENST00000343849.3
ENST00000397368.2
|
CAV3
|
caveolin 3
|
|
chr12_-_57547121
Show fit
|
0.50 |
ENST00000550954.5
ENST00000434715.7
ENST00000678505.1
ENST00000543672.6
ENST00000546670.5
ENST00000548249.6
|
DCTN2
|
dynactin subunit 2
|
|
chr21_-_34887148
Show fit
|
0.50 |
ENST00000399240.5
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr13_-_74133892
Show fit
|
0.49 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12
|
|
chr12_-_12562851
Show fit
|
0.49 |
ENST00000298573.9
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr19_-_50833187
Show fit
|
0.49 |
ENST00000598673.1
|
KLK15
|
kallikrein related peptidase 15
|
|
chr12_-_57547224
Show fit
|
0.48 |
ENST00000678322.1
|
DCTN2
|
dynactin subunit 2
|
|
chr7_-_92836555
Show fit
|
0.48 |
ENST00000424848.3
|
CDK6
|
cyclin dependent kinase 6
|
|
chr10_-_73655984
Show fit
|
0.48 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like
|
|
chr8_+_143734133
Show fit
|
0.47 |
ENST00000527139.7
ENST00000533004.5
|
IQANK1
|
IQ motif and ankyrin repeat containing 1
|
|
chr17_-_7179544
Show fit
|
0.47 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr1_-_153613095
Show fit
|
0.47 |
ENST00000368706.9
|
S100A16
|
S100 calcium binding protein A16
|
|
chr16_+_28823212
Show fit
|
0.47 |
ENST00000570200.5
|
ATXN2L
|
ataxin 2 like
|
|
chr17_-_45425620
Show fit
|
0.47 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr16_+_68737284
Show fit
|
0.46 |
ENST00000261769.10
ENST00000422392.6
|
CDH1
|
cadherin 1
|
|
chr16_+_28822982
Show fit
|
0.46 |
ENST00000336783.9
ENST00000340394.12
ENST00000325215.10
ENST00000382686.8
ENST00000395547.6
ENST00000564304.5
|
ATXN2L
|
ataxin 2 like
|
|
chr19_+_35118456
Show fit
|
0.45 |
ENST00000604621.5
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr8_+_89902529
Show fit
|
0.45 |
ENST00000451899.7
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2
|
|
chr17_-_7179348
Show fit
|
0.45 |
ENST00000573083.1
ENST00000574388.5
ENST00000269299.8
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr22_-_32205001
Show fit
|
0.44 |
ENST00000652607.1
|
RFPL2
|
ret finger protein like 2
|
|
chr2_+_241637612
Show fit
|
0.44 |
ENST00000625810.2
ENST00000402096.5
|
ATG4B
|
autophagy related 4B cysteine peptidase
|
|
chr19_+_54965252
Show fit
|
0.44 |
ENST00000543010.5
ENST00000391721.8
ENST00000339757.11
|
NLRP2
|
NLR family pyrin domain containing 2
|
|
chr19_+_54966343
Show fit
|
0.43 |
ENST00000537859.5
ENST00000585500.5
ENST00000427260.6
ENST00000448584.7
ENST00000263437.10
|
NLRP2
|
NLR family pyrin domain containing 2
|
|
chr9_-_34126661
Show fit
|
0.43 |
ENST00000361264.9
|
DCAF12
|
DDB1 and CUL4 associated factor 12
|
|
chr20_+_836052
Show fit
|
0.43 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A
|
|
chr5_-_78294656
Show fit
|
0.42 |
ENST00000519295.5
ENST00000255194.11
|
AP3B1
|
adaptor related protein complex 3 subunit beta 1
|
|
chr6_+_7107597
Show fit
|
0.42 |
ENST00000379933.7
ENST00000491191.5
ENST00000471433.5
|
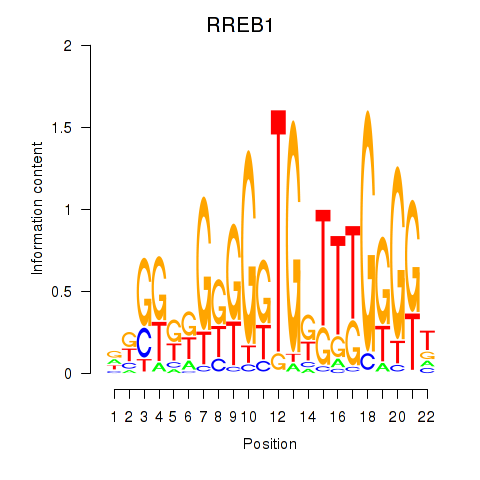
RREB1
|
ras responsive element binding protein 1
|
|
chr11_-_72080389
Show fit
|
0.41 |
ENST00000351960.10
ENST00000541719.5
ENST00000535111.5
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr3_-_179072205
Show fit
|
0.41 |
ENST00000432729.5
|
ZMAT3
|
zinc finger matrin-type 3
|
|
chr12_+_48818478
Show fit
|
0.41 |
ENST00000547818.5
ENST00000301050.7
ENST00000547392.5
|
CACNB3
|
calcium voltage-gated channel auxiliary subunit beta 3
|
|
chr9_-_21994345
Show fit
|
0.41 |
ENST00000579755.2
ENST00000530628.2
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A
|
|
chr3_-_116444983
Show fit
|
0.41 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein
|
|
chr15_+_75206398
Show fit
|
0.41 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr10_+_68332090
Show fit
|
0.40 |
ENST00000354695.5
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3
|
|
chrX_-_152733715
Show fit
|
0.39 |
ENST00000452779.3
ENST00000370287.7
|
CSAG1
|
chondrosarcoma associated gene 1
|
|
chr12_+_118981531
Show fit
|
0.38 |
ENST00000267260.5
|
SRRM4
|
serine/arginine repetitive matrix 4
|
|
chr19_+_48364361
Show fit
|
0.38 |
ENST00000344846.7
|
SYNGR4
|
synaptogyrin 4
|
|
chr6_+_37354046
Show fit
|
0.38 |
ENST00000487950.1
ENST00000469731.5
|
RNF8
|
ring finger protein 8
|
|
chr2_-_136116165
Show fit
|
0.38 |
ENST00000409817.1
|
CXCR4
|
C-X-C motif chemokine receptor 4
|
|
chr11_-_72080472
Show fit
|
0.38 |
ENST00000537217.5
ENST00000366394.7
ENST00000358965.10
ENST00000546131.1
ENST00000393695.8
ENST00000543937.5
ENST00000368959.9
ENST00000541641.5
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr2_+_120735848
Show fit
|
0.38 |
ENST00000361492.9
|
GLI2
|
GLI family zinc finger 2
|
|
chr6_+_112054075
Show fit
|
0.37 |
ENST00000230529.9
ENST00000604763.5
ENST00000368666.7
ENST00000674325.1
ENST00000483439.1
ENST00000409166.5
|
CCN6
|
cellular communication network factor 6
|
|
chr10_-_24723185
Show fit
|
0.37 |
ENST00000376410.7
ENST00000446003.5
|
ARHGAP21
|
Rho GTPase activating protein 21
|
|
chr12_+_65824403
Show fit
|
0.37 |
ENST00000393578.7
ENST00000425208.6
ENST00000536545.5
ENST00000354636.7
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr19_-_15449920
Show fit
|
0.37 |
ENST00000263381.12
ENST00000643092.1
ENST00000673675.1
|
WIZ
|
WIZ zinc finger
|
|
chr19_-_56840661
Show fit
|
0.36 |
ENST00000649735.1
ENST00000593695.5
ENST00000599577.5
ENST00000594389.1
ENST00000648694.1
ENST00000326441.15
ENST00000649680.1
ENST00000649876.1
ENST00000650632.1
ENST00000650102.1
ENST00000647621.1
ENST00000598410.5
ENST00000649233.1
ENST00000593711.6
ENST00000629319.2
ENST00000599935.5
|
PEG3
ZIM2
|
paternally expressed 3
zinc finger imprinted 2
|
|
chr12_+_62260374
Show fit
|
0.36 |
ENST00000312635.10
ENST00000280377.10
ENST00000549237.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chrX_-_152769677
Show fit
|
0.35 |
ENST00000457643.1
ENST00000616035.4
ENST00000412733.1
ENST00000329342.9
|
MAGEA6
|
MAGE family member A6
|
|
chr3_+_57060658
Show fit
|
0.35 |
ENST00000334325.2
|
SPATA12
|
spermatogenesis associated 12
|
|
chr4_+_55853639
Show fit
|
0.35 |
ENST00000381295.7
ENST00000346134.11
ENST00000349598.6
|
EXOC1
|
exocyst complex component 1
|
|
chr16_-_1225257
Show fit
|
0.34 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1
|
|
chr15_+_76336755
Show fit
|
0.34 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2
|
|
chr15_-_77632869
Show fit
|
0.34 |
ENST00000355300.7
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr1_+_209938169
Show fit
|
0.34 |
ENST00000367019.5
ENST00000537238.5
ENST00000637265.1
|
SYT14
|
synaptotagmin 14
|
|
chr17_-_41047267
Show fit
|
0.34 |
ENST00000542137.1
ENST00000391419.3
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr17_+_43847142
Show fit
|
0.33 |
ENST00000377203.8
ENST00000539718.5
ENST00000588884.1
ENST00000293396.12
ENST00000586233.5
|
CD300LG
|
CD300 molecule like family member g
|
|
chr1_+_209938207
Show fit
|
0.33 |
ENST00000472886.5
|
SYT14
|
synaptotagmin 14
|
|
chr8_+_72537219
Show fit
|
0.33 |
ENST00000523207.2
|
KCNB2
|
potassium voltage-gated channel subfamily B member 2
|
|
chr11_+_2301987
Show fit
|
0.33 |
ENST00000612299.4
ENST00000182290.9
|
TSPAN32
|
tetraspanin 32
|
|
chr6_+_37353972
Show fit
|
0.33 |
ENST00000373479.9
|
RNF8
|
ring finger protein 8
|
|
chrX_-_152138547
Show fit
|
0.33 |
ENST00000577437.1
ENST00000427322.6
ENST00000370323.9
ENST00000244096.7
|
MAGEA10-MAGEA5
MAGEA10
|
MAGEA10-MAGEA5 readthrough
MAGE family member A10
|