Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for RUNX3_BCL11A
Z-value: 0.75
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
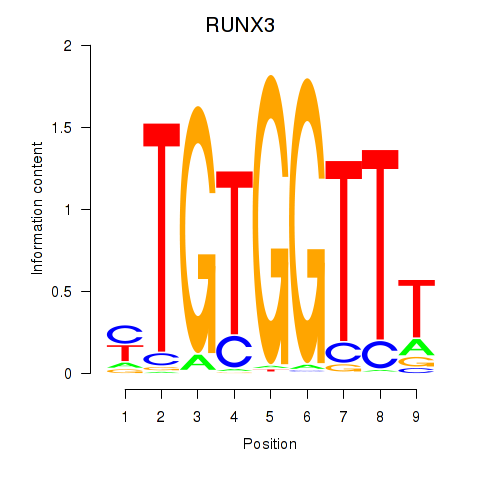
RUNX3
|
ENSG00000020633.19 | RUNX3 |
|
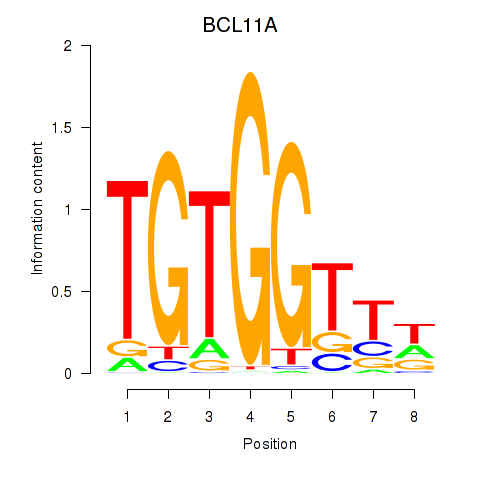
BCL11A
|
ENSG00000119866.22 | BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL11A | hg38_v1_chr2_-_60553409_60553461 | 0.64 | 1.3e-04 | Click! |
| RUNX3 | hg38_v1_chr1_-_24930263_24930282 | 0.41 | 2.5e-02 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_17228215 | 2.02 |
ENST00000544301.7
|
VIM
|
vimentin |
| chr22_+_31082860 | 1.82 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr11_-_58578096 | 1.42 |
ENST00000528954.5
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr3_-_151203201 | 1.25 |
ENST00000480322.1
ENST00000309180.6 |
GPR171
|
G protein-coupled receptor 171 |
| chr3_-_33645253 | 1.19 |
ENST00000333778.10
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_-_102955705 | 1.11 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr5_-_39270623 | 1.06 |
ENST00000512138.1
ENST00000646045.2 |
FYB1
|
FYN binding protein 1 |
| chr10_+_25174969 | 1.05 |
ENST00000376351.4
|
GPR158
|
G protein-coupled receptor 158 |
| chr4_+_87975667 | 1.04 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr10_+_25174796 | 1.04 |
ENST00000650135.1
|
GPR158
|
G protein-coupled receptor 158 |
| chr3_+_100609594 | 1.00 |
ENST00000273352.8
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr4_+_87975829 | 0.97 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr19_-_43780957 | 0.87 |
ENST00000648319.1
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chr13_-_61427849 | 0.83 |
ENST00000409186.1
ENST00000472649.2 |
ENSG00000197991.11
LINC02339
|
novel protein long intergenic non-protein coding RNA 2339 |
| chr9_-_120876356 | 0.83 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr13_-_102401599 | 0.80 |
ENST00000376131.8
|
FGF14
|
fibroblast growth factor 14 |
| chr12_+_14973020 | 0.79 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H |
| chrX_-_107717054 | 0.77 |
ENST00000503515.1
ENST00000372397.6 |
TSC22D3
|
TSC22 domain family member 3 |
| chr9_-_120877026 | 0.74 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19 |
| chr12_+_48978313 | 0.71 |
ENST00000293549.4
|
WNT1
|
Wnt family member 1 |
| chr12_+_48978453 | 0.69 |
ENST00000613114.4
|
WNT1
|
Wnt family member 1 |
| chr10_-_13234329 | 0.69 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr10_-_13234368 | 0.68 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr12_-_51324091 | 0.67 |
ENST00000604560.6
|
BIN2
|
bridging integrator 2 |
| chr1_+_44739825 | 0.66 |
ENST00000372224.9
|
KIF2C
|
kinesin family member 2C |
| chr8_-_71361860 | 0.65 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr8_-_71362054 | 0.65 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr12_-_51324138 | 0.63 |
ENST00000452142.7
|
BIN2
|
bridging integrator 2 |
| chr8_+_22042790 | 0.63 |
ENST00000359441.4
|
FGF17
|
fibroblast growth factor 17 |
| chr1_-_93681829 | 0.63 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr12_-_51324164 | 0.62 |
ENST00000615107.6
|
BIN2
|
bridging integrator 2 |
| chr7_+_30852273 | 0.62 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough |
| chr2_-_157444044 | 0.61 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
| chr10_+_110644306 | 0.59 |
ENST00000369519.4
|
RBM20
|
RNA binding motif protein 20 |
| chr3_-_33645433 | 0.58 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr5_-_39202991 | 0.57 |
ENST00000515010.5
|
FYB1
|
FYN binding protein 1 |
| chr2_-_55419565 | 0.56 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr4_-_57110373 | 0.54 |
ENST00000295666.6
ENST00000514062.2 |
IGFBP7
|
insulin like growth factor binding protein 7 |
| chr1_-_149917826 | 0.53 |
ENST00000369145.1
ENST00000369146.8 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr2_-_55419821 | 0.52 |
ENST00000644630.1
ENST00000471947.2 ENST00000436346.7 ENST00000642200.1 ENST00000413716.7 ENST00000263630.13 ENST00000645072.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr21_+_30396030 | 0.51 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr7_+_120988683 | 0.51 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr11_-_35265696 | 0.47 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_-_24964984 | 0.47 |
ENST00000338888.3
ENST00000399916.5 |
RUNX3
|
RUNX family transcription factor 3 |
| chr1_+_152514474 | 0.45 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1 |
| chr2_-_55296361 | 0.45 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_-_50347710 | 0.43 |
ENST00000342183.9
ENST00000401710.5 |
NRXN1
|
neurexin 1 |
| chr2_-_191150971 | 0.43 |
ENST00000409995.5
ENST00000392320.7 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr1_+_66534082 | 0.42 |
ENST00000683257.1
ENST00000684083.1 ENST00000682938.1 ENST00000683581.1 ENST00000682293.1 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chr19_-_43781249 | 0.42 |
ENST00000615047.4
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4 |
| chrX_-_136780925 | 0.41 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_+_66534107 | 0.41 |
ENST00000371037.9
ENST00000684651.1 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chr11_-_66958366 | 0.40 |
ENST00000651036.1
ENST00000652125.1 ENST00000531614.6 ENST00000524491.6 ENST00000529047.6 ENST00000393960.7 ENST00000393958.7 ENST00000528403.6 ENST00000651854.1 |
PC
|
pyruvate carboxylase |
| chr1_+_66534171 | 0.40 |
ENST00000682762.1
ENST00000424320.6 |
SGIP1
|
SH3GL interacting endocytic adaptor 1 |
| chr4_-_149815826 | 0.40 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr8_+_22042382 | 0.40 |
ENST00000518533.5
|
FGF17
|
fibroblast growth factor 17 |
| chr9_-_92404559 | 0.39 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr11_-_31803620 | 0.39 |
ENST00000639006.1
|
PAX6
|
paired box 6 |
| chr1_+_209686173 | 0.39 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr2_-_2324323 | 0.38 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr6_-_114343012 | 0.38 |
ENST00000312719.10
|
HS3ST5
|
heparan sulfate-glucosamine 3-sulfotransferase 5 |
| chr5_+_35856883 | 0.37 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr7_+_18495723 | 0.37 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr11_-_31803663 | 0.36 |
ENST00000638802.1
ENST00000638878.1 |
PAX6
|
paired box 6 |
| chr4_+_70226116 | 0.36 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr16_+_85908988 | 0.35 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr2_-_2324642 | 0.35 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr13_-_46182136 | 0.34 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr2_+_168802563 | 0.34 |
ENST00000445023.6
|
NOSTRIN
|
nitric oxide synthase trafficking |
| chrX_-_71618455 | 0.34 |
ENST00000373691.4
ENST00000373693.4 |
CXCR3
|
C-X-C motif chemokine receptor 3 |
| chr11_-_19240936 | 0.34 |
ENST00000250024.9
|
E2F8
|
E2F transcription factor 8 |
| chr12_-_70637405 | 0.34 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr1_-_168543990 | 0.33 |
ENST00000367819.3
|
XCL2
|
X-C motif chemokine ligand 2 |
| chr2_+_168802610 | 0.33 |
ENST00000397206.6
ENST00000317647.12 ENST00000397209.6 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr9_-_122213903 | 0.33 |
ENST00000464484.3
|
LHX6
|
LIM homeobox 6 |
| chr5_+_136160986 | 0.33 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr1_+_109910485 | 0.33 |
ENST00000525659.5
|
CSF1
|
colony stimulating factor 1 |
| chr2_+_33134579 | 0.32 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_52618559 | 0.32 |
ENST00000305748.7
|
KRT73
|
keratin 73 |
| chr6_+_73695779 | 0.32 |
ENST00000422508.6
ENST00000437994.6 |
CD109
|
CD109 molecule |
| chr8_-_133060347 | 0.32 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr11_+_124865425 | 0.32 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3 |
| chr6_+_30061231 | 0.32 |
ENST00000376782.6
ENST00000359374.8 ENST00000376785.2 ENST00000332435.10 |
POLR1H
|
RNA polymerase I subunit H |
| chr5_-_139482285 | 0.32 |
ENST00000652110.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chrX_-_15854791 | 0.32 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr9_-_122213874 | 0.31 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr19_-_48993300 | 0.31 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr1_+_109910986 | 0.31 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 |
| chr14_+_85530127 | 0.31 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_-_10684749 | 0.31 |
ENST00000226951.11
|
CLNK
|
cytokine dependent hematopoietic cell linker |
| chr20_+_37777262 | 0.31 |
ENST00000373469.1
|
CTNNBL1
|
catenin beta like 1 |
| chr3_-_71130557 | 0.31 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1 |
| chr3_-_71130963 | 0.30 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr17_-_43758780 | 0.30 |
ENST00000301691.3
|
SOST
|
sclerostin |
| chr6_-_42451261 | 0.30 |
ENST00000372917.8
ENST00000340840.6 ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr13_-_28100556 | 0.30 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr14_-_70188967 | 0.30 |
ENST00000381269.6
ENST00000357887.7 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr19_+_41376692 | 0.30 |
ENST00000447302.6
ENST00000544232.5 ENST00000542945.5 ENST00000540732.3 |
TMEM91
ENSG00000255730.5
|
transmembrane protein 91 novel protein |
| chr14_-_70188937 | 0.29 |
ENST00000356921.7
|
SLC8A3
|
solute carrier family 8 member A3 |
| chr3_+_141386393 | 0.29 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_+_45328203 | 0.28 |
ENST00000371432.7
ENST00000647337.2 ENST00000371438.5 |
RUNX2
|
RUNX family transcription factor 2 |
| chr2_+_181985846 | 0.28 |
ENST00000682840.1
ENST00000409137.7 ENST00000280295.7 |
PPP1R1C
|
protein phosphatase 1 regulatory inhibitor subunit 1C |
| chr3_+_96814552 | 0.28 |
ENST00000470610.6
ENST00000389672.9 |
EPHA6
|
EPH receptor A6 |
| chr5_+_172959511 | 0.28 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26 like 1 |
| chr10_+_5524953 | 0.27 |
ENST00000315238.3
|
CALML3
|
calmodulin like 3 |
| chr14_+_21868822 | 0.27 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr5_-_139482341 | 0.27 |
ENST00000651699.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr12_+_75481204 | 0.27 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1 |
| chr2_+_202073282 | 0.27 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chrX_+_68829009 | 0.27 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr7_-_148883474 | 0.26 |
ENST00000476773.5
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chrX_+_37780049 | 0.26 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chrX_-_15854743 | 0.26 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr12_-_10390023 | 0.26 |
ENST00000240618.11
|
KLRK1
|
killer cell lectin like receptor K1 |
| chr1_-_173205543 | 0.26 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4 |
| chr1_+_6555301 | 0.25 |
ENST00000333172.11
ENST00000351136.7 |
TAS1R1
|
taste 1 receptor member 1 |
| chr1_-_182672232 | 0.25 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr3_+_155083889 | 0.25 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr12_-_16606795 | 0.25 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr4_-_80072993 | 0.25 |
ENST00000681115.1
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr1_-_179143044 | 0.25 |
ENST00000504405.5
ENST00000512653.5 ENST00000344730.7 |
ABL2
|
ABL proto-oncogene 2, non-receptor tyrosine kinase |
| chr3_-_190122317 | 0.24 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr11_-_2301859 | 0.24 |
ENST00000456145.2
ENST00000381153.8 |
C11orf21
|
chromosome 11 open reading frame 21 |
| chr5_-_139482173 | 0.24 |
ENST00000652271.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr17_-_49210567 | 0.24 |
ENST00000507680.6
|
GNGT2
|
G protein subunit gamma transducin 2 |
| chr12_+_79045625 | 0.24 |
ENST00000552744.5
|
SYT1
|
synaptotagmin 1 |
| chr19_-_48321857 | 0.24 |
ENST00000474199.6
|
ODAD1
|
outer dynein arm docking complex subunit 1 |
| chr3_+_155083523 | 0.24 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr17_-_2266131 | 0.24 |
ENST00000570606.5
ENST00000354901.8 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr22_-_23141980 | 0.24 |
ENST00000216036.9
|
RSPH14
|
radial spoke head 14 homolog |
| chr20_+_56412112 | 0.23 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr14_-_34714549 | 0.23 |
ENST00000555765.5
ENST00000672517.1 |
CFL2
|
cofilin 2 |
| chr19_-_48321948 | 0.23 |
ENST00000674294.1
|
ODAD1
|
outer dynein arm docking complex subunit 1 |
| chr1_-_157552470 | 0.22 |
ENST00000361835.8
|
FCRL5
|
Fc receptor like 5 |
| chr9_+_116153783 | 0.22 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1 |
| chr14_-_34714579 | 0.22 |
ENST00000298159.11
|
CFL2
|
cofilin 2 |
| chr19_+_7348930 | 0.22 |
ENST00000668164.2
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor 18 |
| chr4_-_151226427 | 0.22 |
ENST00000304527.8
ENST00000409598.8 |
SH3D19
|
SH3 domain containing 19 |
| chr14_+_85530163 | 0.22 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_-_58417547 | 0.22 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43 |
| chr20_+_56412393 | 0.22 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr2_+_37344594 | 0.21 |
ENST00000404976.5
ENST00000338415.8 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr17_-_58417521 | 0.21 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr2_+_33134620 | 0.21 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_49154433 | 0.21 |
ENST00000454032.5
ENST00000304421.8 |
FSHR
|
follicle stimulating hormone receptor |
| chr11_-_62689523 | 0.21 |
ENST00000317449.5
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr6_-_52244500 | 0.21 |
ENST00000336123.5
|
IL17F
|
interleukin 17F |
| chr2_-_49154507 | 0.21 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor |
| chr1_-_24143112 | 0.21 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1 |
| chr10_+_99659430 | 0.20 |
ENST00000370489.5
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr6_-_29045175 | 0.20 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr2_+_74529923 | 0.20 |
ENST00000258080.8
ENST00000352222.7 |
HTRA2
|
HtrA serine peptidase 2 |
| chr15_+_43185382 | 0.20 |
ENST00000300213.9
|
CCNDBP1
|
cyclin D1 binding protein 1 |
| chr17_-_10114546 | 0.20 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7 |
| chr3_-_71130892 | 0.20 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr1_+_236394268 | 0.19 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain |
| chr20_+_56412249 | 0.19 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr14_-_59484317 | 0.19 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr22_+_39901075 | 0.19 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr3_+_133124807 | 0.19 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108 |
| chr11_+_124611420 | 0.19 |
ENST00000284288.3
|
PANX3
|
pannexin 3 |
| chr12_-_68159732 | 0.19 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
| chr16_+_87602478 | 0.19 |
ENST00000284262.3
|
JPH3
|
junctophilin 3 |
| chr9_-_34048868 | 0.19 |
ENST00000379239.9
ENST00000684158.1 ENST00000379238.7 ENST00000360802.6 |
UBAP2
|
ubiquitin associated protein 2 |
| chrX_+_79144664 | 0.18 |
ENST00000645147.2
|
GPR174
|
G protein-coupled receptor 174 |
| chr7_-_29195186 | 0.18 |
ENST00000449801.5
ENST00000409850.5 |
CPVL
|
carboxypeptidase vitellogenic like |
| chr19_-_50365625 | 0.18 |
ENST00000598915.5
ENST00000253719.7 |
NAPSA
|
napsin A aspartic peptidase |
| chr7_-_87475647 | 0.18 |
ENST00000649586.2
ENST00000265723.8 |
ABCB4
|
ATP binding cassette subfamily B member 4 |
| chr11_+_77179750 | 0.18 |
ENST00000458169.2
|
MYO7A
|
myosin VIIA |
| chr20_+_31440626 | 0.18 |
ENST00000376309.4
|
DEFB123
|
defensin beta 123 |
| chr19_+_40751179 | 0.18 |
ENST00000243563.8
ENST00000601393.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr5_+_55102635 | 0.17 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr14_+_64214136 | 0.17 |
ENST00000557084.1
ENST00000458046.6 |
SYNE2
|
spectrin repeat containing nuclear envelope protein 2 |
| chr6_+_25279061 | 0.17 |
ENST00000461945.1
|
CARMIL1
|
capping protein regulator and myosin 1 linker 1 |
| chr2_+_183078736 | 0.17 |
ENST00000354221.5
|
DUSP19
|
dual specificity phosphatase 19 |
| chr12_-_54300974 | 0.17 |
ENST00000435572.7
ENST00000553070.5 |
NFE2
|
nuclear factor, erythroid 2 |
| chr6_+_31575557 | 0.17 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor |
| chr16_+_31355165 | 0.17 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr3_-_185821092 | 0.17 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr15_+_49423233 | 0.16 |
ENST00000560270.1
ENST00000267843.9 ENST00000560979.1 |
FGF7
|
fibroblast growth factor 7 |
| chr7_+_142598016 | 0.16 |
ENST00000620773.1
|
TRBV16
|
T cell receptor beta variable 16 |
| chr2_+_202073249 | 0.16 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr1_+_109910892 | 0.16 |
ENST00000369802.7
ENST00000420111.6 |
CSF1
|
colony stimulating factor 1 |
| chr6_-_34556319 | 0.16 |
ENST00000374037.8
ENST00000544425.2 |
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr19_-_14674886 | 0.16 |
ENST00000344373.8
ENST00000595472.1 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr5_-_128339191 | 0.16 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chrX_-_77969638 | 0.15 |
ENST00000458128.3
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr17_+_8310814 | 0.15 |
ENST00000579439.5
ENST00000421050.2 |
ARHGEF15
|
Rho guanine nucleotide exchange factor 15 |
| chr1_+_39081316 | 0.15 |
ENST00000484793.5
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr2_+_102418642 | 0.15 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr19_+_39997031 | 0.15 |
ENST00000599504.5
ENST00000596894.5 ENST00000601138.5 ENST00000347077.9 ENST00000600094.5 |
ZNF546
|
zinc finger protein 546 |
| chr6_-_107459504 | 0.15 |
ENST00000369037.9
ENST00000369031.4 |
PDSS2
|
decaprenyl diphosphate synthase subunit 2 |
| chr19_-_14674829 | 0.15 |
ENST00000443157.6
ENST00000253673.6 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr11_+_60971777 | 0.15 |
ENST00000542157.5
ENST00000433107.6 ENST00000352009.9 ENST00000452451.6 |
CD6
|
CD6 molecule |
| chr19_-_409134 | 0.15 |
ENST00000332235.8
|
C2CD4C
|
C2 calcium dependent domain containing 4C |
| chr6_-_118710065 | 0.15 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr17_+_49210699 | 0.14 |
ENST00000225941.6
|
ABI3
|
ABI family member 3 |
| chr17_-_41811930 | 0.14 |
ENST00000393928.6
|
P3H4
|
prolyl 3-hydroxylase family member 4 (inactive) |
| chr11_-_31804067 | 0.14 |
ENST00000639548.1
ENST00000640125.1 ENST00000481563.6 ENST00000639079.1 ENST00000638762.1 ENST00000638346.1 |
PAX6
|
paired box 6 |
| chr19_-_42132465 | 0.14 |
ENST00000529067.5
ENST00000529952.5 ENST00000342301.8 ENST00000389341.9 |
POU2F2
|
POU class 2 homeobox 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.5 | 1.4 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.3 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.8 | GO:0060611 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 1.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 1.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.4 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 2.0 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.4 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.4 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 1.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.4 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:2001025 | response to cyclosporin A(GO:1905237) positive regulation of response to drug(GO:2001025) |
| 0.1 | 0.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 1.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.5 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 1.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 1.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.8 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.6 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.6 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.3 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.7 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 1.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 2.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.4 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 1.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.8 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.3 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.5 | GO:0097109 | acetylcholine receptor binding(GO:0033130) neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 2.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |