Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
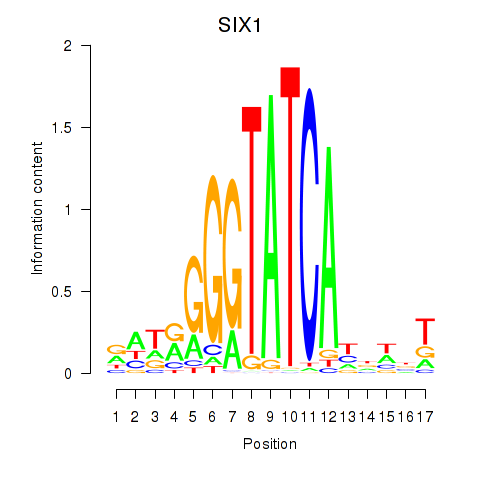
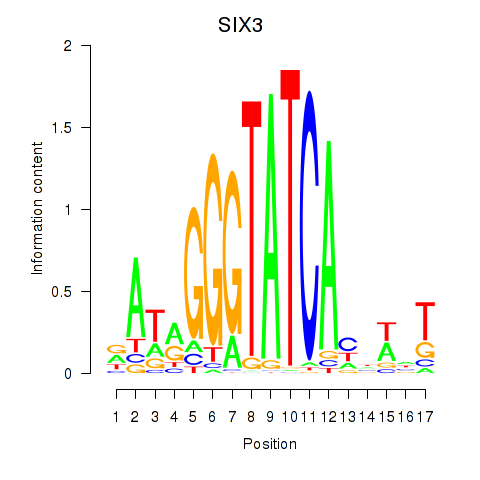
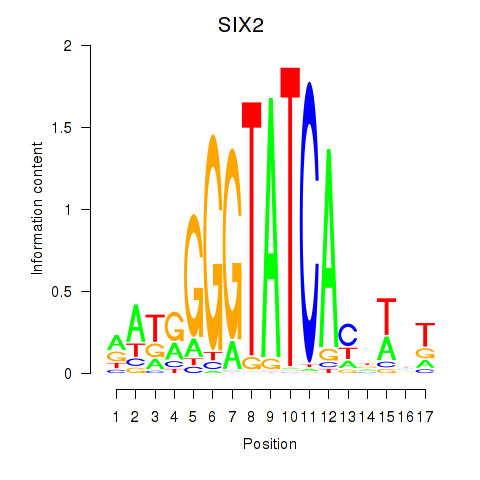
Results for SIX1_SIX3_SIX2
Z-value: 0.46
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.12 | SIX1 |
|
SIX3
|
ENSG00000138083.5 | SIX3 |
|
SIX2
|
ENSG00000170577.8 | SIX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX3 | hg38_v1_chr2_+_44941695_44941708 | 0.41 | 2.6e-02 | Click! |
| SIX2 | hg38_v1_chr2_-_45009401_45009457 | 0.17 | 3.6e-01 | Click! |
| SIX1 | hg38_v1_chr14_-_60649449_60649490 | 0.10 | 6.0e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_56525925 | 0.79 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr1_-_108200849 | 0.75 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr6_+_84033717 | 0.65 |
ENST00000257776.5
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr11_-_58731968 | 0.61 |
ENST00000278400.3
|
GLYAT
|
glycine-N-acyltransferase |
| chr1_-_206946448 | 0.53 |
ENST00000356495.5
|
PIGR
|
polymeric immunoglobulin receptor |
| chr13_-_46105009 | 0.49 |
ENST00000439329.5
ENST00000674625.1 ENST00000181383.10 |
CPB2
|
carboxypeptidase B2 |
| chr14_+_22271921 | 0.46 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr14_-_24442241 | 0.45 |
ENST00000555355.5
ENST00000553343.5 ENST00000556523.1 ENST00000556249.1 ENST00000538105.6 ENST00000555225.5 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U member 1 |
| chr1_+_26410809 | 0.40 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr11_-_32430811 | 0.38 |
ENST00000379079.8
ENST00000530998.5 |
WT1
|
WT1 transcription factor |
| chr11_+_72189528 | 0.37 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chrX_-_66040107 | 0.37 |
ENST00000455586.6
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr6_-_75493773 | 0.37 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1 |
| chrX_-_66040057 | 0.37 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr9_+_12693327 | 0.35 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chrX_-_66040072 | 0.35 |
ENST00000374737.9
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr3_+_119147375 | 0.32 |
ENST00000490594.2
|
ENSG00000251012.2
|
novel chromosome 3 open reading frame 30 (C3orf30) and uroplakin 1B (UPK1B) |
| chr1_+_45500287 | 0.32 |
ENST00000401061.9
ENST00000616135.1 |
MMACHC
|
metabolism of cobalamin associated C |
| chr10_-_46046264 | 0.30 |
ENST00000581478.5
ENST00000582163.3 |
MSMB
|
microseminoprotein beta |
| chr1_+_241532121 | 0.29 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr7_-_99971845 | 0.29 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr19_+_735026 | 0.28 |
ENST00000592155.5
ENST00000590161.2 |
PALM
|
paralemmin |
| chr11_-_58731936 | 0.28 |
ENST00000344743.8
|
GLYAT
|
glycine-N-acyltransferase |
| chr2_+_148978361 | 0.28 |
ENST00000678720.1
ENST00000678856.1 ENST00000677080.1 |
KIF5C
|
kinesin family member 5C |
| chr1_-_89198868 | 0.28 |
ENST00000355754.7
|
GBP4
|
guanylate binding protein 4 |
| chr11_-_34511710 | 0.28 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr19_+_11346556 | 0.26 |
ENST00000587531.5
|
CCDC159
|
coiled-coil domain containing 159 |
| chr5_+_126423363 | 0.26 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr5_+_126423122 | 0.26 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr3_-_151461011 | 0.25 |
ENST00000282466.4
|
IGSF10
|
immunoglobulin superfamily member 10 |
| chr1_-_169367746 | 0.24 |
ENST00000367811.8
ENST00000472647.5 |
NME7
|
NME/NM23 family member 7 |
| chr6_-_24935942 | 0.24 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_182450132 | 0.24 |
ENST00000294854.13
|
RGSL1
|
regulator of G protein signaling like 1 |
| chr11_+_72080313 | 0.24 |
ENST00000307198.11
ENST00000538413.6 ENST00000642648.1 ENST00000289488.7 |
ENSG00000284922.2
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chr6_-_46921926 | 0.23 |
ENST00000283296.12
|
ADGRF5
|
adhesion G protein-coupled receptor F5 |
| chr1_+_14929734 | 0.23 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr5_+_126423176 | 0.23 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr2_-_206765274 | 0.22 |
ENST00000454776.6
ENST00000449792.5 ENST00000374412.8 |
MDH1B
|
malate dehydrogenase 1B |
| chr1_+_13254212 | 0.22 |
ENST00000622421.2
|
PRAMEF5
|
PRAME family member 5 |
| chr3_-_19934189 | 0.20 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B |
| chr3_+_149474688 | 0.20 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_+_11346499 | 0.19 |
ENST00000458408.6
ENST00000586451.5 ENST00000588592.5 |
CCDC159
|
coiled-coil domain containing 159 |
| chr10_+_7703300 | 0.19 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr2_+_131527833 | 0.19 |
ENST00000295171.10
ENST00000467992.6 ENST00000409856.8 |
CCDC74A
|
coiled-coil domain containing 74A |
| chr1_+_160739239 | 0.18 |
ENST00000368043.8
|
SLAMF7
|
SLAM family member 7 |
| chr21_+_25639272 | 0.18 |
ENST00000400532.5
ENST00000312957.9 |
JAM2
|
junctional adhesion molecule 2 |
| chr14_+_50560137 | 0.18 |
ENST00000358385.12
|
ATL1
|
atlastin GTPase 1 |
| chr1_-_12947580 | 0.17 |
ENST00000376189.5
|
PRAMEF6
|
PRAME family member 6 |
| chr3_-_11643871 | 0.17 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr22_+_45284941 | 0.17 |
ENST00000216211.9
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr20_+_36236433 | 0.17 |
ENST00000397286.7
ENST00000679519.1 ENST00000679667.1 ENST00000680933.1 ENST00000680247.1 ENST00000320849.9 ENST00000680811.1 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor |
| chr12_-_14951106 | 0.16 |
ENST00000541644.5
ENST00000545895.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr6_-_154430495 | 0.16 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr10_+_112375196 | 0.16 |
ENST00000393081.6
|
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr8_+_75539862 | 0.16 |
ENST00000396423.4
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr1_+_160739265 | 0.16 |
ENST00000368042.7
|
SLAMF7
|
SLAM family member 7 |
| chr7_+_77122609 | 0.16 |
ENST00000285871.5
|
CCDC146
|
coiled-coil domain containing 146 |
| chr2_-_222320124 | 0.16 |
ENST00000678139.1
|
ENSG00000288658.1
|
novel protein, ortholog of Gm2102 (M. musculus) |
| chr8_+_75539893 | 0.15 |
ENST00000674002.1
|
HNF4G
|
hepatocyte nuclear factor 4 gamma |
| chr11_-_128842467 | 0.15 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr2_+_218672027 | 0.15 |
ENST00000392105.7
ENST00000455724.5 ENST00000295709.8 |
STK36
|
serine/threonine kinase 36 |
| chr5_-_35195236 | 0.15 |
ENST00000509839.5
|
PRLR
|
prolactin receptor |
| chr6_+_33075952 | 0.14 |
ENST00000418931.7
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr10_+_7703340 | 0.14 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_+_55262152 | 0.13 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr19_+_55947832 | 0.13 |
ENST00000291971.7
ENST00000590542.1 |
NLRP8
|
NLR family pyrin domain containing 8 |
| chr3_+_69763549 | 0.13 |
ENST00000472437.5
|
MITF
|
melanocyte inducing transcription factor |
| chr4_+_88378733 | 0.13 |
ENST00000273960.7
ENST00000380265.9 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chrX_-_72875974 | 0.13 |
ENST00000595412.5
|
DMRTC1
|
DMRT like family C1 |
| chr2_+_233917371 | 0.13 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr21_+_25639251 | 0.13 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr3_+_44712634 | 0.13 |
ENST00000449836.5
ENST00000296091.8 ENST00000436624.7 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr2_+_234050732 | 0.13 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr1_+_13068677 | 0.13 |
ENST00000614839.4
|
PRAMEF25
|
PRAME family member 25 |
| chr6_-_149896059 | 0.13 |
ENST00000532335.5
|
RAET1E
|
retinoic acid early transcript 1E |
| chr15_+_62561361 | 0.13 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr3_-_42875871 | 0.12 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr12_+_48727431 | 0.12 |
ENST00000548380.6
|
TEX49
|
testis expressed 49 |
| chrX_+_115289709 | 0.12 |
ENST00000371920.4
ENST00000371921.5 |
LUZP4
|
leucine zipper protein 4 |
| chr7_-_44159278 | 0.12 |
ENST00000345378.7
|
GCK
|
glucokinase |
| chr9_-_34895722 | 0.12 |
ENST00000603640.6
ENST00000603592.1 ENST00000340783.11 |
FAM205C
|
family with sequence similarity 205 member C |
| chr11_+_61752603 | 0.11 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr4_+_88378842 | 0.11 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_-_43082692 | 0.11 |
ENST00000406487.6
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr19_-_57935353 | 0.11 |
ENST00000595569.1
ENST00000599852.1 ENST00000425570.7 ENST00000601593.5 ENST00000396147.6 |
ZNF418
|
zinc finger protein 418 |
| chr3_-_46464868 | 0.11 |
ENST00000417439.5
ENST00000231751.9 ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr2_-_158380960 | 0.11 |
ENST00000409187.5
|
CCDC148
|
coiled-coil domain containing 148 |
| chr6_+_31586859 | 0.11 |
ENST00000433492.5
|
LST1
|
leukocyte specific transcript 1 |
| chr5_+_161685748 | 0.11 |
ENST00000523217.5
|
GABRA6
|
gamma-aminobutyric acid type A receptor subunit alpha6 |
| chr10_-_101843920 | 0.11 |
ENST00000358038.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr11_+_36296281 | 0.11 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr15_+_81134257 | 0.10 |
ENST00000286732.5
|
CFAP161
|
cilia and flagella associated protein 161 |
| chr10_-_112446932 | 0.10 |
ENST00000682195.1
ENST00000683505.1 ENST00000683895.1 ENST00000682839.1 ENST00000682055.1 |
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr12_-_89526253 | 0.10 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr10_-_101843765 | 0.10 |
ENST00000370046.5
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr19_-_54063905 | 0.10 |
ENST00000645936.1
ENST00000376626.5 ENST00000366170.6 ENST00000425006.3 |
VSTM1
|
V-set and transmembrane domain containing 1 |
| chr11_-_126062782 | 0.10 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr3_+_23945271 | 0.10 |
ENST00000312521.9
|
NR1D2
|
nuclear receptor subfamily 1 group D member 2 |
| chr8_+_117135259 | 0.10 |
ENST00000519688.5
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr16_+_14071303 | 0.10 |
ENST00000571589.6
ENST00000574998.5 ENST00000574045.5 |
MRTFB
|
myocardin related transcription factor B |
| chr1_+_40477248 | 0.10 |
ENST00000372706.6
|
ZFP69
|
ZFP69 zinc finger protein |
| chrX_+_13569593 | 0.09 |
ENST00000361306.6
ENST00000380602.3 |
EGFL6
|
EGF like domain multiple 6 |
| chr16_+_75222609 | 0.09 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr16_+_21684429 | 0.09 |
ENST00000388956.8
|
OTOA
|
otoancorin |
| chr1_-_167935987 | 0.09 |
ENST00000367846.8
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr17_-_74531467 | 0.09 |
ENST00000314401.3
ENST00000392621.6 |
CD300LB
|
CD300 molecule like family member b |
| chr21_-_34511243 | 0.09 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr1_+_12857086 | 0.09 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr2_-_89297785 | 0.09 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr15_+_75258599 | 0.09 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family member C |
| chr11_-_118565992 | 0.09 |
ENST00000264020.6
|
IFT46
|
intraflagellar transport 46 |
| chr19_+_46297032 | 0.09 |
ENST00000377670.9
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chr1_-_56819365 | 0.09 |
ENST00000343433.7
|
FYB2
|
FYN binding protein 2 |
| chr1_-_183418364 | 0.09 |
ENST00000287713.7
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr17_+_4771878 | 0.09 |
ENST00000270560.4
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr3_+_111998915 | 0.08 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr1_-_12831410 | 0.08 |
ENST00000619922.1
|
PRAMEF11
|
PRAME family member 11 |
| chr21_-_42350987 | 0.08 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2 |
| chr5_+_140175205 | 0.08 |
ENST00000644078.1
|
CYSTM1
|
cysteine rich transmembrane module containing 1 |
| chr8_-_27605271 | 0.08 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr3_+_111999189 | 0.08 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr14_+_22304051 | 0.08 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr3_+_111999326 | 0.08 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr5_-_132737518 | 0.08 |
ENST00000403231.6
ENST00000378735.5 ENST00000618515.4 ENST00000378746.8 |
KIF3A
|
kinesin family member 3A |
| chr16_-_1954682 | 0.08 |
ENST00000268661.8
|
RPL3L
|
ribosomal protein L3 like |
| chr1_-_12945416 | 0.08 |
ENST00000415464.6
|
PRAMEF6
|
PRAME family member 6 |
| chr12_+_20695553 | 0.08 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr2_-_219170023 | 0.08 |
ENST00000409878.8
ENST00000455516.6 ENST00000295738.11 |
SLC23A3
|
solute carrier family 23 member 3 |
| chr2_-_55269038 | 0.08 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr11_+_64555956 | 0.08 |
ENST00000377581.7
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr5_-_161548124 | 0.08 |
ENST00000520240.5
ENST00000517901.5 ENST00000353437.10 |
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr11_-_44950839 | 0.07 |
ENST00000395648.7
ENST00000531928.6 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr1_+_119507203 | 0.07 |
ENST00000369413.8
ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr8_+_93754844 | 0.07 |
ENST00000684064.1
ENST00000498673.5 ENST00000518319.5 |
TMEM67
|
transmembrane protein 67 |
| chr12_-_54984667 | 0.07 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr4_-_155376876 | 0.07 |
ENST00000433024.5
ENST00000379248.6 |
MAP9
|
microtubule associated protein 9 |
| chr1_+_154321107 | 0.07 |
ENST00000484864.1
|
AQP10
|
aquaporin 10 |
| chr11_-_49208589 | 0.07 |
ENST00000256999.7
|
FOLH1
|
folate hydrolase 1 |
| chr9_+_35909481 | 0.07 |
ENST00000443779.3
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr5_-_146182475 | 0.07 |
ENST00000674158.1
ENST00000674191.1 ENST00000274562.13 |
LARS1
|
leucyl-tRNA synthetase 1 |
| chr11_-_44950867 | 0.07 |
ENST00000528290.5
ENST00000525680.6 ENST00000530035.5 ENST00000527685.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr1_-_207911027 | 0.07 |
ENST00000310833.12
|
CD34
|
CD34 molecule |
| chr9_-_72060605 | 0.07 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr1_-_13155961 | 0.07 |
ENST00000624207.1
|
PRAMEF26
|
PRAME family member 26 |
| chr3_+_42091316 | 0.07 |
ENST00000327628.10
|
TRAK1
|
trafficking kinesin protein 1 |
| chr17_+_78187317 | 0.07 |
ENST00000409257.9
ENST00000591256.5 ENST00000589256.5 ENST00000588800.5 ENST00000591952.5 ENST00000327898.9 ENST00000586542.5 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr12_+_133181409 | 0.07 |
ENST00000416488.5
ENST00000228289.9 ENST00000541211.6 ENST00000536435.7 ENST00000500625.7 ENST00000539248.6 ENST00000542711.6 ENST00000536899.6 ENST00000542986.6 ENST00000611984.4 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr5_-_146182591 | 0.07 |
ENST00000510191.5
ENST00000674277.1 ENST00000674447.1 ENST00000674270.1 ENST00000394434.7 ENST00000674290.1 ENST00000674398.1 ENST00000674174.1 |
LARS1
|
leucyl-tRNA synthetase 1 |
| chr22_+_36860973 | 0.07 |
ENST00000447071.5
ENST00000397147.7 ENST00000248899.11 |
NCF4
|
neutrophil cytosolic factor 4 |
| chr12_-_108633864 | 0.07 |
ENST00000550948.2
|
SELPLG
|
selectin P ligand |
| chr19_-_3600581 | 0.07 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr1_+_154321076 | 0.07 |
ENST00000324978.8
|
AQP10
|
aquaporin 10 |
| chr3_+_111998739 | 0.07 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr1_+_12791397 | 0.06 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr6_+_150368997 | 0.06 |
ENST00000392255.7
ENST00000500320.7 ENST00000344419.8 |
IYD
|
iodotyrosine deiodinase |
| chr5_-_82278341 | 0.06 |
ENST00000510210.5
ENST00000512493.5 ENST00000296674.13 ENST00000507980.1 ENST00000511844.1 ENST00000651545.1 |
RPS23
|
ribosomal protein S23 |
| chr16_-_81181312 | 0.06 |
ENST00000527937.1
ENST00000531391.5 |
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr2_+_89884740 | 0.06 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr9_-_72060590 | 0.06 |
ENST00000652156.1
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr14_+_58244821 | 0.06 |
ENST00000216455.9
ENST00000412908.6 ENST00000557508.5 |
PSMA3
|
proteasome 20S subunit alpha 3 |
| chr1_+_207104226 | 0.06 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr10_-_112446891 | 0.06 |
ENST00000369404.3
ENST00000369405.7 ENST00000626395.2 |
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr2_-_157875820 | 0.06 |
ENST00000672582.1
ENST00000673324.1 ENST00000539637.6 ENST00000413751.1 ENST00000424669.6 ENST00000684348.1 |
ACVR1
|
activin A receptor type 1 |
| chr20_-_32657362 | 0.06 |
ENST00000360785.6
|
C20orf203
|
chromosome 20 open reading frame 203 |
| chr6_-_32192845 | 0.06 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr2_+_105337515 | 0.06 |
ENST00000410049.1
ENST00000258457.7 |
C2orf49
|
chromosome 2 open reading frame 49 |
| chr1_-_207911384 | 0.06 |
ENST00000356522.4
|
CD34
|
CD34 molecule |
| chr1_-_159076742 | 0.06 |
ENST00000368130.9
|
AIM2
|
absent in melanoma 2 |
| chr10_+_46375721 | 0.06 |
ENST00000616785.1
ENST00000611655.1 ENST00000619162.5 |
ENSG00000285402.1
ANXA8L1
|
novel transcript annexin A8 like 1 |
| chr12_-_89526164 | 0.06 |
ENST00000548729.5
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr10_-_112446734 | 0.06 |
ENST00000684507.1
|
ZDHHC6
|
zinc finger DHHC-type palmitoyltransferase 6 |
| chr9_-_76692181 | 0.06 |
ENST00000376717.6
ENST00000223609.10 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr4_-_121381007 | 0.06 |
ENST00000394427.3
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr11_+_85628573 | 0.06 |
ENST00000393375.5
ENST00000358867.11 ENST00000534341.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr4_-_155376935 | 0.06 |
ENST00000311277.9
|
MAP9
|
microtubule associated protein 9 |
| chr3_+_178536205 | 0.06 |
ENST00000420517.6
|
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr6_+_31615984 | 0.06 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr2_-_89213917 | 0.06 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr1_-_230745574 | 0.06 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr12_-_6689450 | 0.06 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr11_-_10568571 | 0.06 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr1_+_160151597 | 0.06 |
ENST00000368081.9
|
ATP1A4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr10_-_101843777 | 0.06 |
ENST00000356640.7
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2 |
| chr19_-_10928512 | 0.06 |
ENST00000588347.5
|
YIPF2
|
Yip1 domain family member 2 |
| chr22_+_20080211 | 0.05 |
ENST00000383024.6
ENST00000351989.8 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chr1_+_27725945 | 0.05 |
ENST00000373954.11
ENST00000419687.6 |
FAM76A
|
family with sequence similarity 76 member A |
| chr4_-_155376797 | 0.05 |
ENST00000650955.1
ENST00000515654.5 |
MAP9
|
microtubule associated protein 9 |
| chr9_+_35910076 | 0.05 |
ENST00000636776.1
|
SPAAR
|
small regulatory polypeptide of amino acid response |
| chr19_-_38229714 | 0.05 |
ENST00000416611.5
|
DPF1
|
double PHD fingers 1 |
| chr11_+_64555924 | 0.05 |
ENST00000301891.9
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr3_-_142000353 | 0.05 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr5_+_112976757 | 0.05 |
ENST00000389063.3
|
DCP2
|
decapping mRNA 2 |
| chr6_-_24358036 | 0.05 |
ENST00000378454.8
|
DCDC2
|
doublecortin domain containing 2 |
| chrX_+_102651366 | 0.05 |
ENST00000415986.5
ENST00000444152.5 ENST00000361600.9 |
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr19_+_55605639 | 0.05 |
ENST00000568956.2
|
ZNF865
|
zinc finger protein 865 |
| chr16_+_84768246 | 0.05 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr19_+_57584131 | 0.05 |
ENST00000536878.6
ENST00000597219.1 ENST00000598689.1 ENST00000597850.2 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr19_+_7497544 | 0.05 |
ENST00000361664.7
|
TEX45
|
testis expressed 45 |
| chr11_+_64555684 | 0.05 |
ENST00000377585.7
|
SLC22A11
|
solute carrier family 22 member 11 |
| chr9_-_122159742 | 0.05 |
ENST00000373768.4
|
NDUFA8
|
NADH:ubiquinone oxidoreductase subunit A8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.5 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 0.4 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:2001107 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.1 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.3 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.4 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.0 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.0 | GO:1902996 | regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.7 | GO:0097009 | energy homeostasis(GO:0097009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.9 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |