Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for SIX5_SMARCC2_HCFC1
Z-value: 2.09
Transcription factors associated with SIX5_SMARCC2_HCFC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
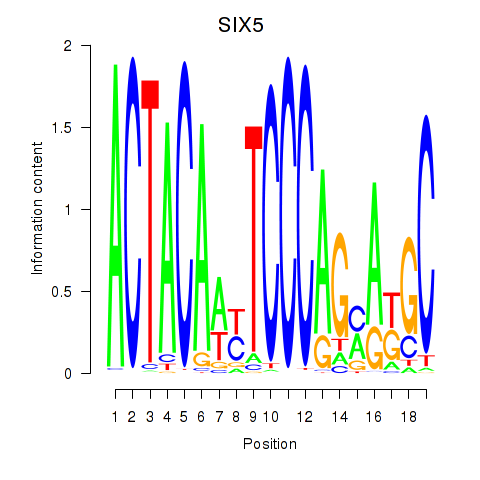
SIX5
|
ENSG00000177045.10 | SIX5 |
|
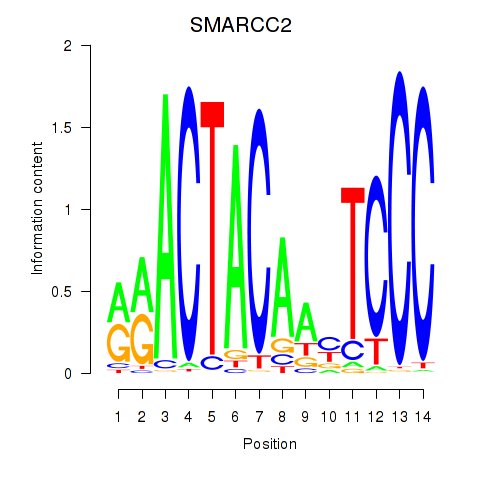
SMARCC2
|
ENSG00000139613.12 | SMARCC2 |
|
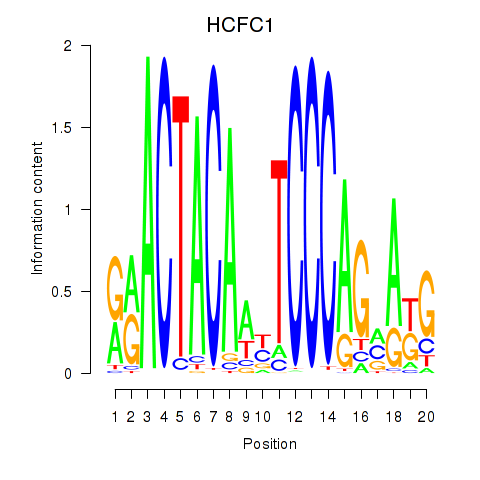
HCFC1
|
ENSG00000172534.14 | HCFC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMARCC2 | hg38_v1_chr12_-_56189548_56189609 | 0.41 | 2.4e-02 | Click! |
| HCFC1 | hg38_v1_chrX_-_153971169_153971233, hg38_v1_chrX_-_153971810_153971844 | 0.28 | 1.4e-01 | Click! |
| SIX5 | hg38_v1_chr19_-_45768843_45768869 | -0.25 | 1.9e-01 | Click! |
Activity profile of SIX5_SMARCC2_HCFC1 motif
Sorted Z-values of SIX5_SMARCC2_HCFC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX5_SMARCC2_HCFC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 1.6 | 4.7 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.2 | 5.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 1.2 | 3.7 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 1.2 | 3.7 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 1.1 | 8.0 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 1.0 | 3.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.8 | 5.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.8 | 3.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.8 | 2.3 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.7 | 6.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.7 | 2.9 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.6 | 2.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.6 | 2.3 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.5 | 1.6 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.5 | 2.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.5 | 7.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.5 | 10.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.5 | 2.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 9.5 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.5 | 1.5 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.5 | 1.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.5 | 3.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.4 | 3.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 1.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.4 | 1.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.4 | 3.9 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 1.2 | GO:0031445 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) |
| 0.4 | 10.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.4 | 2.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 1.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.4 | 0.7 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 | 1.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 2.0 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.3 | 4.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 3.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.3 | 1.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.3 | 1.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 2.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 5.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 1.9 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 0.8 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.3 | 3.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.3 | 1.3 | GO:1900114 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.2 | 8.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.2 | 1.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.2 | 0.5 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.2 | 1.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 6.1 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 4.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.7 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.2 | 1.3 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.2 | 1.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 2.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 4.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.6 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.2 | 2.0 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.2 | 1.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.8 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 0.6 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 0.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 0.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.9 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 0.7 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 1.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.2 | 5.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 1.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.5 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 0.8 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 2.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 0.5 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 0.6 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 2.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 1.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.7 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 1.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 2.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.5 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 2.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.9 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 6.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.5 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.8 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 6.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.6 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 1.6 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 0.9 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 1.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 1.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.7 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.4 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 2.9 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.5 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 7.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.5 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.3 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 3.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.5 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.6 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.6 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 2.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0035562 | regulation of chromatin binding(GO:0035561) negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 6.8 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 3.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.4 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 1.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.9 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.6 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.9 | GO:0060340 | MDA-5 signaling pathway(GO:0039530) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.7 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) response to hypobaric hypoxia(GO:1990910) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 3.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 4.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0071335 | submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 1.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 1.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 3.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 1.9 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.0 | 1.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.4 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 1.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 1.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 1.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0093001 | glycolytic process through glucose-1-phosphate(GO:0061622) glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.7 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 1.7 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 2.6 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0042791 | tRNA transcription(GO:0009304) 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.4 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 1.3 | 6.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.7 | 7.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.6 | 10.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.6 | 4.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 2.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.6 | 4.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 2.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 3.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 1.2 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.4 | 1.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.4 | 2.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 1.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 4.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.8 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 8.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 1.0 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.3 | 8.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 10.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 1.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 3.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.2 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 1.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.2 | 0.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 2.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.8 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 1.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 2.0 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 0.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 0.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.9 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 4.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 1.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 3.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 1.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.8 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 10.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 2.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 4.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 6.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 2.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 4.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 2.2 | 6.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.6 | 4.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.4 | 4.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 1.2 | 3.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.1 | 9.6 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.9 | 4.5 | GO:0004803 | transposase activity(GO:0004803) |
| 0.7 | 2.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.7 | 5.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.6 | 7.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 8.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 3.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 2.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 5.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 1.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.4 | 3.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 2.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.4 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 2.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.0 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.3 | 2.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 1.0 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.3 | 2.5 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.3 | 0.9 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 1.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 0.7 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 2.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 1.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.7 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.2 | 7.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 1.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 2.6 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 1.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 5.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 1.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 12.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.8 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 1.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 1.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.7 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.6 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.6 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.5 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 1.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.6 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.5 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.4 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.8 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 1.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 3.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.2 | GO:0004577 | N-acetylglucosaminyldiphosphodolichol N-acetylglucosaminyltransferase activity(GO:0004577) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 2.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 5.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 1.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 4.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 1.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 67.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.1 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) apolipoprotein receptor binding(GO:0034190) apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 3.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 7.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 4.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 3.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 5.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 48.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 8.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 7.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.5 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 3.8 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |