Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
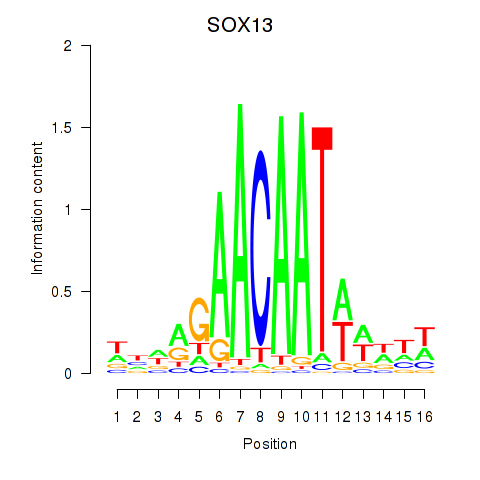
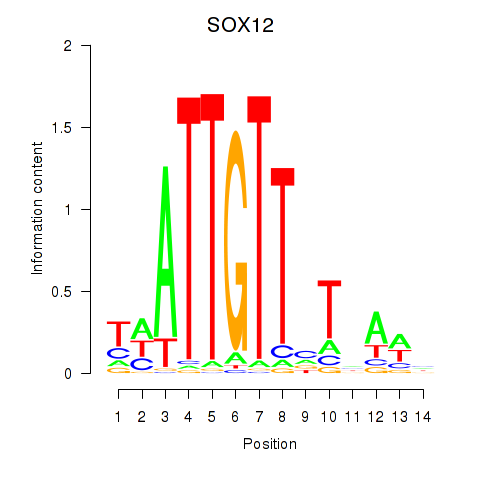
Results for SOX13_SOX12
Z-value: 1.09
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.15 | SOX13 |
|
SOX12
|
ENSG00000177732.9 | SOX12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg38_v1_chr20_+_325536_325565 | -0.24 | 2.1e-01 | Click! |
| SOX13 | hg38_v1_chr1_+_204073104_204073121 | -0.00 | 9.9e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91111460 | 7.16 |
ENST00000266718.5
|
LUM
|
lumican |
| chr10_+_116427839 | 5.67 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr2_+_102311502 | 3.34 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr1_-_153460644 | 2.71 |
ENST00000368723.4
|
S100A7
|
S100 calcium binding protein A7 |
| chr4_-_56681288 | 2.57 |
ENST00000556376.6
ENST00000420433.6 |
HOPX
|
HOP homeobox |
| chr19_+_20923275 | 2.14 |
ENST00000300540.7
ENST00000595854.5 ENST00000601284.5 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr4_-_68245683 | 2.09 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr3_+_186640355 | 1.83 |
ENST00000382134.7
ENST00000265029.8 |
FETUB
|
fetuin B |
| chr12_-_10130241 | 1.73 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr6_-_49744378 | 1.70 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr6_-_49744434 | 1.69 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr11_-_55936400 | 1.58 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr1_+_151060357 | 1.55 |
ENST00000368921.5
|
MLLT11
|
MLLT11 transcription factor 7 cofactor |
| chrX_-_15600953 | 1.47 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr4_+_186227501 | 1.46 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr5_-_147831663 | 1.46 |
ENST00000296695.10
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr7_+_101127095 | 1.39 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr3_+_186640411 | 1.39 |
ENST00000382136.3
|
FETUB
|
fetuin B |
| chr1_+_153031195 | 1.37 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr2_+_33436304 | 1.26 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr6_-_25830557 | 1.19 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr6_+_32637419 | 1.18 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr1_-_153041111 | 1.18 |
ENST00000360379.4
|
SPRR2D
|
small proline rich protein 2D |
| chr12_-_8662703 | 1.17 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr1_-_153057504 | 1.15 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A |
| chr1_+_153416517 | 1.13 |
ENST00000368729.9
|
S100A7A
|
S100 calcium binding protein A7A |
| chr8_-_7416863 | 1.13 |
ENST00000318157.3
|
DEFB4B
|
defensin beta 4B |
| chr18_+_49562049 | 1.12 |
ENST00000261292.9
ENST00000427224.6 ENST00000580036.5 |
LIPG
|
lipase G, endothelial type |
| chr4_-_158173004 | 1.08 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr5_-_147831627 | 1.06 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor Kazal type 1 |
| chr15_+_85380625 | 1.05 |
ENST00000560302.5
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr15_-_89211803 | 1.04 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr8_-_13276491 | 1.01 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr19_+_47713412 | 1.01 |
ENST00000538399.1
ENST00000263277.8 |
EHD2
|
EH domain containing 2 |
| chr2_-_216013517 | 1.00 |
ENST00000263268.11
|
MREG
|
melanoregulin |
| chr12_-_8662619 | 1.00 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr22_-_18936142 | 0.96 |
ENST00000438924.5
ENST00000457083.1 ENST00000357068.11 ENST00000420436.5 ENST00000334029.6 ENST00000610940.4 |
PRODH
|
proline dehydrogenase 1 |
| chr6_+_12290353 | 0.94 |
ENST00000379375.6
|
EDN1
|
endothelin 1 |
| chr1_+_74235377 | 0.93 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_+_9961165 | 0.92 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1 |
| chr19_+_21082140 | 0.91 |
ENST00000616183.1
ENST00000596053.5 |
ZNF714
|
zinc finger protein 714 |
| chr2_+_157257687 | 0.91 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr1_+_64203610 | 0.91 |
ENST00000371077.10
ENST00000611228.4 |
UBE2U
|
ubiquitin conjugating enzyme E2 U |
| chr17_-_40703744 | 0.90 |
ENST00000264651.3
|
KRT24
|
keratin 24 |
| chr7_+_77538027 | 0.88 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr1_-_152325232 | 0.88 |
ENST00000368799.2
|
FLG
|
filaggrin |
| chr21_-_31160904 | 0.88 |
ENST00000636887.1
|
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chrX_+_71283186 | 0.82 |
ENST00000535149.5
|
NONO
|
non-POU domain containing octamer binding |
| chrX_+_136197020 | 0.80 |
ENST00000370676.7
|
FHL1
|
four and a half LIM domains 1 |
| chr10_-_104085847 | 0.80 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain |
| chr19_+_15949008 | 0.80 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor family 10 subfamily H member 4 |
| chr4_+_70197924 | 0.77 |
ENST00000514097.5
|
ODAM
|
odontogenic, ameloblast associated |
| chr19_+_41708635 | 0.77 |
ENST00000617332.4
ENST00000615021.4 ENST00000616453.1 ENST00000405816.5 ENST00000435837.2 |
CEACAM5
ENSG00000267881.2
|
CEA cell adhesion molecule 5 novel protein, readthrough between CEACAM5-CEACAM6 |
| chr3_-_132684685 | 0.77 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr2_-_171066936 | 0.73 |
ENST00000453628.1
ENST00000434911.6 |
TLK1
|
tousled like kinase 1 |
| chr14_+_56117702 | 0.72 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_+_86547070 | 0.72 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr19_+_41376499 | 0.71 |
ENST00000392002.7
|
TMEM91
|
transmembrane protein 91 |
| chr19_-_23687163 | 0.70 |
ENST00000601010.5
ENST00000601935.5 ENST00000600313.5 ENST00000596211.5 ENST00000359788.9 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr14_-_74875998 | 0.70 |
ENST00000556489.4
ENST00000673765.1 |
PROX2
|
prospero homeobox 2 |
| chr4_-_158173042 | 0.69 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr10_-_96358989 | 0.67 |
ENST00000371172.8
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr2_-_40453438 | 0.66 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr2_+_90172802 | 0.65 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr19_+_41708585 | 0.65 |
ENST00000398599.8
ENST00000221992.11 |
CEACAM5
|
CEA cell adhesion molecule 5 |
| chr18_+_63587297 | 0.64 |
ENST00000269489.9
|
SERPINB13
|
serpin family B member 13 |
| chr11_+_34632464 | 0.63 |
ENST00000531794.5
|
EHF
|
ETS homologous factor |
| chr5_+_90640718 | 0.63 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr6_-_132659178 | 0.62 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr22_-_50085414 | 0.62 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr12_-_8650529 | 0.61 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5 |
| chr2_-_216013582 | 0.61 |
ENST00000620139.4
|
MREG
|
melanoregulin |
| chr11_-_48983826 | 0.61 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr12_+_21527017 | 0.61 |
ENST00000535033.5
|
SPX
|
spexin hormone |
| chr17_-_676348 | 0.61 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr12_-_7444139 | 0.61 |
ENST00000416109.2
ENST00000313599.8 |
CD163L1
|
CD163 molecule like 1 |
| chrX_+_136197039 | 0.59 |
ENST00000370683.6
|
FHL1
|
four and a half LIM domains 1 |
| chr2_+_57907621 | 0.59 |
ENST00000648897.1
ENST00000435505.6 ENST00000417641.6 |
VRK2
|
VRK serine/threonine kinase 2 |
| chr18_+_63587336 | 0.59 |
ENST00000344731.10
|
SERPINB13
|
serpin family B member 13 |
| chr22_-_50085331 | 0.59 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr20_+_142573 | 0.57 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126 |
| chr10_+_84452208 | 0.57 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr12_-_11395556 | 0.56 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr21_+_42199686 | 0.55 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chrX_+_30243715 | 0.55 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr11_-_18236795 | 0.55 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr8_+_24294044 | 0.53 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_+_140855882 | 0.53 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr12_-_8662073 | 0.53 |
ENST00000535411.5
ENST00000540087.5 |
MFAP5
|
microfibril associated protein 5 |
| chr19_-_14674829 | 0.52 |
ENST00000443157.6
ENST00000253673.6 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr8_-_63026179 | 0.51 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase |
| chr17_+_4788926 | 0.51 |
ENST00000331264.8
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr6_-_130970428 | 0.51 |
ENST00000529208.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr11_+_57597563 | 0.50 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr12_+_8843236 | 0.50 |
ENST00000541459.5
|
A2ML1
|
alpha-2-macroglobulin like 1 |
| chr20_-_23751256 | 0.50 |
ENST00000398402.1
|
CST1
|
cystatin SN |
| chr14_-_50561119 | 0.50 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr15_-_79090760 | 0.50 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr12_-_84892120 | 0.50 |
ENST00000680379.1
|
SLC6A15
|
solute carrier family 6 member 15 |
| chr3_+_111998739 | 0.47 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr13_-_37598750 | 0.47 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr19_-_14674886 | 0.47 |
ENST00000344373.8
ENST00000595472.1 |
ADGRE3
|
adhesion G protein-coupled receptor E3 |
| chr5_-_22853320 | 0.47 |
ENST00000504376.6
ENST00000382254.6 |
CDH12
|
cadherin 12 |
| chr2_-_89085787 | 0.46 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr9_+_92964272 | 0.46 |
ENST00000468206.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_75206044 | 0.46 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr2_+_69013414 | 0.45 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr1_+_24319342 | 0.45 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr11_+_35180279 | 0.45 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_111245725 | 0.45 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr6_+_106086316 | 0.44 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr16_-_10694922 | 0.44 |
ENST00000283025.7
|
TEKT5
|
tektin 5 |
| chr19_-_8981342 | 0.44 |
ENST00000397910.8
|
MUC16
|
mucin 16, cell surface associated |
| chr2_-_89245596 | 0.43 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr10_+_116545907 | 0.43 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr2_+_209580024 | 0.43 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr14_+_22147988 | 0.43 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr2_-_89027700 | 0.43 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr18_+_26133843 | 0.43 |
ENST00000415576.7
ENST00000343848.10 ENST00000308268.10 |
PSMA8
|
proteasome 20S subunit alpha 8 |
| chr7_+_134891566 | 0.42 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr6_+_26204552 | 0.42 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5 |
| chr5_+_141421020 | 0.42 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_-_203182805 | 0.42 |
ENST00000404436.2
|
CHI3L1
|
chitinase 3 like 1 |
| chr5_-_95682968 | 0.42 |
ENST00000274432.13
|
SPATA9
|
spermatogenesis associated 9 |
| chr1_-_89022827 | 0.42 |
ENST00000370481.9
ENST00000564665.1 |
GBP3
|
guanylate binding protein 3 |
| chr14_+_73239599 | 0.42 |
ENST00000554301.5
ENST00000555445.5 |
PAPLN
|
papilin, proteoglycan like sulfated glycoprotein |
| chr19_-_8896090 | 0.41 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr2_+_90114838 | 0.41 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chrX_-_154371210 | 0.41 |
ENST00000369856.8
ENST00000422373.6 ENST00000360319.9 |
FLNA
|
filamin A |
| chr6_+_26183750 | 0.41 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr1_+_76867469 | 0.41 |
ENST00000477717.6
|
ST6GALNAC5
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr3_-_197226351 | 0.41 |
ENST00000656428.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr5_-_77492309 | 0.41 |
ENST00000296679.9
ENST00000507029.5 |
WDR41
|
WD repeat domain 41 |
| chr1_+_21551260 | 0.41 |
ENST00000374832.5
|
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr14_-_94129577 | 0.40 |
ENST00000238609.4
|
IFI27L2
|
interferon alpha inducible protein 27 like 2 |
| chr12_+_50842920 | 0.40 |
ENST00000551456.5
ENST00000398458.4 |
TMPRSS12
|
transmembrane serine protease 12 |
| chr20_+_3786772 | 0.40 |
ENST00000344256.10
ENST00000379598.9 |
CDC25B
|
cell division cycle 25B |
| chr1_+_171185293 | 0.39 |
ENST00000209929.10
|
FMO2
|
flavin containing dimethylaniline monoxygenase 2 |
| chr12_-_8662808 | 0.39 |
ENST00000359478.7
ENST00000396549.6 |
MFAP5
|
microfibril associated protein 5 |
| chr15_-_34318761 | 0.39 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr11_-_10808304 | 0.38 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr17_+_55751021 | 0.38 |
ENST00000268896.10
ENST00000576183.5 ENST00000573500.5 |
PCTP
|
phosphatidylcholine transfer protein |
| chr2_+_102418642 | 0.38 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr18_+_63775369 | 0.38 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr17_-_74776323 | 0.38 |
ENST00000582870.5
ENST00000581136.5 ENST00000579218.5 ENST00000583476.5 ENST00000580301.5 ENST00000583757.5 ENST00000357814.8 ENST00000582524.5 |
NAT9
|
N-acetyltransferase 9 (putative) |
| chr16_+_4495852 | 0.38 |
ENST00000575129.5
ENST00000398595.7 ENST00000414777.5 |
HMOX2
|
heme oxygenase 2 |
| chr11_-_60183011 | 0.38 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr3_-_139539577 | 0.37 |
ENST00000619087.4
|
RBP1
|
retinol binding protein 1 |
| chr11_+_35176575 | 0.37 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_102317450 | 0.37 |
ENST00000615299.4
ENST00000527309.2 ENST00000526421.6 ENST00000263464.9 |
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr4_+_52862308 | 0.36 |
ENST00000248706.5
|
RASL11B
|
RAS like family 11 member B |
| chr4_-_24912954 | 0.36 |
ENST00000502801.1
ENST00000635206.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr12_+_75334655 | 0.36 |
ENST00000378695.9
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr21_-_32813695 | 0.36 |
ENST00000479548.2
ENST00000490358.5 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr2_-_227379297 | 0.36 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr14_+_22508602 | 0.36 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr7_-_41703062 | 0.36 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr8_+_24294107 | 0.35 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr22_-_35840218 | 0.35 |
ENST00000414461.6
ENST00000416721.6 ENST00000449924.6 ENST00000262829.11 ENST00000397305.3 |
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr6_+_27824084 | 0.35 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr12_-_10986912 | 0.35 |
ENST00000506868.1
|
TAS2R50
|
taste 2 receptor member 50 |
| chr15_-_64989894 | 0.35 |
ENST00000416889.6
ENST00000204566.7 |
SPG21
|
SPG21 abhydrolase domain containing, maspardin |
| chr3_+_57890011 | 0.35 |
ENST00000494088.6
ENST00000438794.5 |
SLMAP
|
sarcolemma associated protein |
| chr10_-_28334439 | 0.35 |
ENST00000441595.2
|
MPP7
|
membrane palmitoylated protein 7 |
| chr11_+_7088991 | 0.34 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr12_-_91153149 | 0.34 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_-_4786354 | 0.34 |
ENST00000328739.6
ENST00000441199.2 ENST00000416307.6 |
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr10_-_125816596 | 0.34 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr6_+_36871841 | 0.34 |
ENST00000359359.6
|
C6orf89
|
chromosome 6 open reading frame 89 |
| chr14_+_94110728 | 0.34 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr15_+_67138001 | 0.34 |
ENST00000439724.7
|
SMAD3
|
SMAD family member 3 |
| chrX_-_15601077 | 0.34 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_-_89217339 | 0.34 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr12_+_75334675 | 0.33 |
ENST00000312442.2
|
GLIPR1L1
|
GLIPR1 like 1 |
| chr13_-_49413749 | 0.33 |
ENST00000610540.4
ENST00000347776.9 ENST00000409082.5 |
CAB39L
|
calcium binding protein 39 like |
| chr10_-_125816510 | 0.33 |
ENST00000650587.1
|
UROS
|
uroporphyrinogen III synthase |
| chr12_+_59664677 | 0.33 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr12_+_68809002 | 0.33 |
ENST00000539479.6
ENST00000393415.7 ENST00000523991.5 ENST00000543323.5 ENST00000393416.7 |
MDM2
|
MDM2 proto-oncogene |
| chr11_+_59787067 | 0.33 |
ENST00000528805.1
|
STX3
|
syntaxin 3 |
| chr15_-_79971164 | 0.33 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr17_-_7394800 | 0.33 |
ENST00000574401.5
|
PLSCR3
|
phospholipid scramblase 3 |
| chr20_-_57711536 | 0.32 |
ENST00000265626.8
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_+_113058637 | 0.32 |
ENST00000346807.7
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr1_+_84181630 | 0.32 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_-_33473884 | 0.32 |
ENST00000297990.9
ENST00000353159.6 ENST00000379471.3 |
NOL6
|
nucleolar protein 6 |
| chr8_+_104223320 | 0.32 |
ENST00000339750.3
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_107803215 | 0.32 |
ENST00000340010.10
ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 member 3 |
| chr4_+_99511008 | 0.32 |
ENST00000514652.5
ENST00000326581.9 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr18_-_12656716 | 0.32 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr12_+_101594849 | 0.32 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr4_+_155758990 | 0.32 |
ENST00000505154.5
ENST00000652626.1 ENST00000502959.5 ENST00000264424.13 ENST00000505764.5 ENST00000507146.5 ENST00000503520.5 |
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_+_116672187 | 0.32 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr19_-_55140922 | 0.32 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr15_+_21651844 | 0.31 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr1_+_207752046 | 0.31 |
ENST00000367042.6
ENST00000322875.8 ENST00000322918.9 ENST00000354848.5 ENST00000357714.5 ENST00000358170.6 ENST00000367041.5 ENST00000367047.5 ENST00000360212.6 ENST00000480003.5 |
CD46
|
CD46 molecule |
| chr10_-_107164692 | 0.31 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1 |
| chr14_+_22281097 | 0.31 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr11_-_64917200 | 0.31 |
ENST00000377264.8
ENST00000421419.3 |
ATG2A
|
autophagy related 2A |
| chr8_+_103819244 | 0.31 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_53377749 | 0.31 |
ENST00000507166.5
|
ENSG00000282278.1
|
novel FIP1L1-PDGFRA fusion protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.5 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.6 | 3.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.5 | 1.4 | GO:0001300 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.5 | 3.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.4 | 1.8 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.3 | 1.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 1.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 0.9 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.3 | 1.8 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 1.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.3 | 0.6 | GO:2000170 | regulation of nitric oxide mediated signal transduction(GO:0010749) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 2.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.2 | 0.9 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 0.6 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.2 | 0.5 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 | 0.7 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 0.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.4 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.4 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.4 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.4 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.4 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.3 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.1 | 0.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.9 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 1.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 2.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 1.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 4.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 5.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.9 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.6 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 1.2 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.9 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.7 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.3 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 1.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.5 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.2 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 2.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 2.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.1 | 1.8 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.2 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.9 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.5 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.3 | GO:0019323 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.0 | 0.4 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 1.4 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:2000230 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.4 | GO:0070424 | negative regulation of necroptotic process(GO:0060546) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 1.2 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 2.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 2.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:1903537 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 0.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 0.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 3.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.5 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.3 | GO:0033593 | BRCA2-MAGE-D1 complex(GO:0033593) |
| 0.1 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 4.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 4.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.0 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.4 | 1.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 7.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 1.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 0.5 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 2.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 3.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.1 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.1 | 0.3 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.1 | 11.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.5 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.7 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.3 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 2.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 6.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.6 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 1.6 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 3.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 3.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 2.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 2.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |