Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
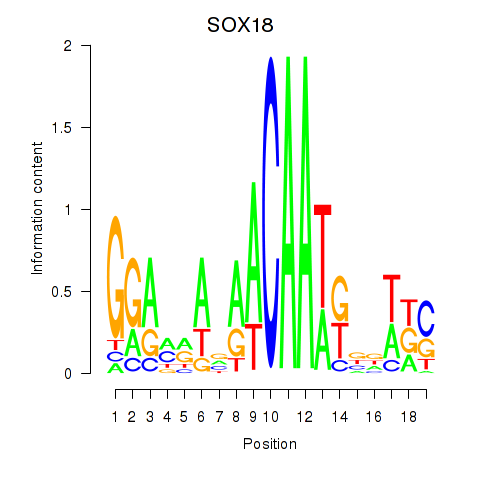
Results for SOX18
Z-value: 0.98
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.7 | SOX18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg38_v1_chr20_-_64049631_64049646 | -0.04 | 8.5e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_171185293 | 5.92 |
ENST00000209929.10
|
FMO2
|
flavin containing dimethylaniline monoxygenase 2 |
| chr22_+_31092447 | 4.94 |
ENST00000455608.5
|
SMTN
|
smoothelin |
| chr5_-_137499293 | 4.79 |
ENST00000510689.5
ENST00000394945.6 |
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 |
| chr15_-_74212219 | 3.01 |
ENST00000449139.6
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr15_-_74212256 | 2.84 |
ENST00000416286.7
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr9_-_96302142 | 2.72 |
ENST00000648799.1
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr9_-_96302104 | 2.40 |
ENST00000375262.4
ENST00000650386.1 |
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr13_+_77535681 | 2.35 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535742 | 2.33 |
ENST00000377246.7
|
SCEL
|
sciellin |
| chr13_+_77535669 | 2.33 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr6_-_27912396 | 2.21 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr16_+_66603874 | 2.19 |
ENST00000563672.5
ENST00000424011.6 |
CMTM3
|
CKLF like MARVEL transmembrane domain containing 3 |
| chr9_-_96302170 | 2.18 |
ENST00000375263.8
|
HSD17B3
|
hydroxysteroid 17-beta dehydrogenase 3 |
| chr1_-_158554405 | 2.14 |
ENST00000641282.1
ENST00000641622.1 |
OR6Y1
|
olfactory receptor family 6 subfamily Y member 1 |
| chr19_+_39655907 | 2.13 |
ENST00000392051.4
|
LGALS16
|
galectin 16 |
| chr4_-_67883987 | 1.91 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D |
| chr12_-_2876986 | 1.85 |
ENST00000342628.6
ENST00000361953.7 |
FOXM1
|
forkhead box M1 |
| chr19_-_7040179 | 1.83 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr19_+_7049321 | 1.82 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2 |
| chr12_+_15322529 | 1.77 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr2_+_32946944 | 1.73 |
ENST00000404816.7
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_76570544 | 1.62 |
ENST00000640006.1
|
ENSG00000284526.1
|
novel protein |
| chr19_+_7030578 | 1.56 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr2_+_26786020 | 1.55 |
ENST00000335756.9
ENST00000233505.12 |
CENPA
|
centromere protein A |
| chr22_+_44026283 | 1.53 |
ENST00000619710.4
|
PARVB
|
parvin beta |
| chr12_-_2877113 | 1.53 |
ENST00000627656.2
ENST00000359843.8 |
FOXM1
|
forkhead box M1 |
| chr1_-_209784521 | 1.50 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr12_-_8662703 | 1.49 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr19_-_7058640 | 1.48 |
ENST00000333843.8
|
MBD3L3
|
methyl-CpG binding domain protein 3 like 3 |
| chr8_+_7539627 | 1.47 |
ENST00000533250.2
|
PRR23D1
|
proline rich 23 domain containing 1 |
| chrX_-_49184789 | 1.46 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chrX_-_132961390 | 1.37 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr2_-_210303608 | 1.37 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr15_+_21651844 | 1.33 |
ENST00000623441.1
|
OR4N4C
|
olfactory receptor family 4 subfamily N member 4C |
| chr6_-_111793871 | 1.26 |
ENST00000368667.6
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr2_+_209653171 | 1.21 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr3_+_156674579 | 1.20 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr19_+_40751179 | 1.15 |
ENST00000243563.8
ENST00000601393.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr1_+_153031195 | 1.14 |
ENST00000307098.5
|
SPRR1B
|
small proline rich protein 1B |
| chr7_+_142511614 | 1.10 |
ENST00000390377.1
|
TRBV7-7
|
T cell receptor beta variable 7-7 |
| chrX_+_136536099 | 1.08 |
ENST00000440515.5
ENST00000456412.1 |
VGLL1
|
vestigial like family member 1 |
| chr3_+_160677152 | 1.08 |
ENST00000320767.4
|
ARL14
|
ADP ribosylation factor like GTPase 14 |
| chr9_+_33240159 | 1.06 |
ENST00000379721.4
|
SPINK4
|
serine peptidase inhibitor Kazal type 4 |
| chr13_-_33205997 | 1.04 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr7_+_134866831 | 1.04 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chrX_+_3066833 | 1.03 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr17_-_17281232 | 1.00 |
ENST00000417352.5
ENST00000268717.10 |
COPS3
|
COP9 signalosome subunit 3 |
| chr19_-_7021431 | 0.99 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr13_-_44161257 | 0.98 |
ENST00000400419.2
|
SMIM2
|
small integral membrane protein 2 |
| chr3_-_53844617 | 0.98 |
ENST00000481668.5
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr15_+_67125707 | 0.98 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr4_+_153152163 | 0.96 |
ENST00000676423.1
ENST00000675745.1 ENST00000676348.1 ENST00000676408.1 ENST00000674874.1 ENST00000675315.1 ENST00000675518.1 |
TRIM2
ENSG00000288637.1
|
tripartite motif containing 2 novel protein |
| chr3_+_19148500 | 0.95 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr1_-_153608136 | 0.95 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16 |
| chr16_-_90019821 | 0.95 |
ENST00000568838.2
|
DBNDD1
|
dysbindin domain containing 1 |
| chr3_+_124384513 | 0.94 |
ENST00000682540.1
ENST00000522553.6 ENST00000682695.1 ENST00000682674.1 ENST00000684382.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr1_-_155910881 | 0.93 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr12_-_7936177 | 0.93 |
ENST00000544291.1
ENST00000075120.12 |
SLC2A3
|
solute carrier family 2 member 3 |
| chr1_-_113887375 | 0.92 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2 like 15 |
| chr3_+_133746385 | 0.89 |
ENST00000482271.5
ENST00000402696.9 |
TF
|
transferrin |
| chr2_-_70553440 | 0.89 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr15_-_29968864 | 0.89 |
ENST00000356107.11
|
TJP1
|
tight junction protein 1 |
| chr15_+_22094522 | 0.88 |
ENST00000328795.5
|
OR4N4
|
olfactory receptor family 4 subfamily N member 4 |
| chr2_+_74654228 | 0.87 |
ENST00000611975.4
ENST00000357877.7 ENST00000339773.9 ENST00000434486.5 |
SEMA4F
|
ssemaphorin 4F |
| chr12_-_11092313 | 0.87 |
ENST00000531678.1
|
TAS2R43
|
taste 2 receptor member 43 |
| chr13_-_46182136 | 0.86 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr9_-_72060605 | 0.86 |
ENST00000377024.8
ENST00000651200.2 ENST00000652752.1 |
C9orf57
|
chromosome 9 open reading frame 57 |
| chr13_-_46142834 | 0.86 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr19_-_50639827 | 0.86 |
ENST00000593901.5
ENST00000600079.6 |
SYT3
|
synaptotagmin 3 |
| chr16_-_90019414 | 0.84 |
ENST00000002501.11
|
DBNDD1
|
dysbindin domain containing 1 |
| chr3_+_136957948 | 0.84 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr4_-_76023489 | 0.84 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr1_+_64203610 | 0.84 |
ENST00000371077.10
ENST00000611228.4 |
UBE2U
|
ubiquitin conjugating enzyme E2 U |
| chr4_+_153466324 | 0.82 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chr13_+_31739520 | 0.82 |
ENST00000298386.7
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr15_+_90930173 | 0.81 |
ENST00000480470.5
ENST00000394275.7 |
UNC45A
|
unc-45 myosin chaperone A |
| chr3_+_111732474 | 0.80 |
ENST00000393923.7
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr12_-_10986912 | 0.79 |
ENST00000506868.1
|
TAS2R50
|
taste 2 receptor member 50 |
| chr4_-_122456725 | 0.77 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr11_-_119095456 | 0.76 |
ENST00000530167.1
|
H2AX
|
H2A.X variant histone |
| chr2_+_100974849 | 0.75 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr15_-_55249029 | 0.75 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr4_-_173399102 | 0.74 |
ENST00000296506.8
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr14_+_94110728 | 0.74 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr11_-_34357994 | 0.73 |
ENST00000435224.3
|
ABTB2
|
ankyrin repeat and BTB domain containing 2 |
| chr3_+_124384757 | 0.72 |
ENST00000684374.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr3_+_102435015 | 0.70 |
ENST00000306176.5
ENST00000466937.2 |
ZPLD1
|
zona pellucida like domain containing 1 |
| chr6_-_31806937 | 0.70 |
ENST00000375661.6
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_+_57756230 | 0.70 |
ENST00000295951.7
ENST00000659705.1 ENST00000671191.1 |
SLMAP
|
sarcolemma associated protein |
| chr11_-_7941708 | 0.69 |
ENST00000642047.1
|
OR10A3
|
olfactory receptor family 10 subfamily A member 3 |
| chr5_-_79512794 | 0.69 |
ENST00000282260.10
ENST00000508576.5 ENST00000535690.1 |
HOMER1
|
homer scaffold protein 1 |
| chr17_+_41255384 | 0.69 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr12_-_11395556 | 0.69 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr3_+_111998739 | 0.68 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr12_+_95858928 | 0.68 |
ENST00000266735.9
ENST00000553192.5 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr15_-_64825608 | 0.68 |
ENST00000559239.2
ENST00000268043.8 ENST00000333425.10 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chrX_+_86714623 | 0.68 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr17_-_75855204 | 0.66 |
ENST00000589642.5
ENST00000593002.1 ENST00000590221.5 ENST00000587374.5 ENST00000585462.5 ENST00000254806.8 ENST00000433525.6 ENST00000626827.2 |
WBP2
|
WW domain binding protein 2 |
| chr9_-_21351378 | 0.66 |
ENST00000380210.1
|
IFNA6
|
interferon alpha 6 |
| chr7_+_157336988 | 0.66 |
ENST00000262177.9
ENST00000417758.5 ENST00000443280.5 |
DNAJB6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr3_+_183265302 | 0.65 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr13_+_31739542 | 0.65 |
ENST00000380314.2
|
RXFP2
|
relaxin family peptide receptor 2 |
| chr1_-_113887574 | 0.64 |
ENST00000393316.8
|
BCL2L15
|
BCL2 like 15 |
| chr15_+_62066975 | 0.63 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chr2_-_36966503 | 0.62 |
ENST00000263918.9
|
STRN
|
striatin |
| chrX_+_47218670 | 0.62 |
ENST00000357227.9
ENST00000519758.5 ENST00000520893.5 ENST00000622098.4 ENST00000517426.5 |
CDK16
|
cyclin dependent kinase 16 |
| chr19_+_4402615 | 0.61 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
| chr11_-_118176576 | 0.60 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2 |
| chr8_-_85341705 | 0.60 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr12_-_10409757 | 0.59 |
ENST00000309384.2
|
KLRC4
|
killer cell lectin like receptor C4 |
| chr4_-_113979635 | 0.59 |
ENST00000315366.8
|
ARSJ
|
arylsulfatase family member J |
| chr6_+_25652272 | 0.58 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr5_-_102498913 | 0.58 |
ENST00000513675.1
ENST00000379807.7 ENST00000506729.6 |
SLCO6A1
|
solute carrier organic anion transporter family member 6A1 |
| chr1_-_13116854 | 0.56 |
ENST00000621994.3
|
HNRNPCL2
|
heterogeneous nuclear ribonucleoprotein C like 2 |
| chr1_-_201154418 | 0.56 |
ENST00000435310.5
ENST00000485839.6 |
TMEM9
|
transmembrane protein 9 |
| chr11_+_89926762 | 0.56 |
ENST00000526396.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr10_+_84424919 | 0.55 |
ENST00000543283.2
ENST00000494586.5 |
CCSER2
|
coiled-coil serine rich protein 2 |
| chr1_-_201154459 | 0.55 |
ENST00000414605.2
ENST00000367330.6 ENST00000367334.9 ENST00000367332.5 |
TMEM9
|
transmembrane protein 9 |
| chr11_-_102625332 | 0.55 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr7_+_142111739 | 0.55 |
ENST00000550469.6
ENST00000477922.3 |
MGAM2
|
maltase-glucoamylase 2 (putative) |
| chr5_-_43515128 | 0.55 |
ENST00000306862.7
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr1_-_155911365 | 0.54 |
ENST00000651833.1
ENST00000539040.5 ENST00000651853.1 |
RIT1
|
Ras like without CAAX 1 |
| chrX_+_47193796 | 0.54 |
ENST00000442035.5
ENST00000335972.11 ENST00000457753.5 |
UBA1
|
ubiquitin like modifier activating enzyme 1 |
| chr19_-_41688167 | 0.52 |
ENST00000602225.1
|
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr14_-_22469673 | 0.52 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr8_-_4994972 | 0.52 |
ENST00000520002.5
ENST00000602557.5 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr11_+_65182413 | 0.51 |
ENST00000531068.5
ENST00000527699.5 ENST00000533909.5 ENST00000527323.5 |
CAPN1
|
calpain 1 |
| chr22_+_26621952 | 0.50 |
ENST00000354760.4
|
CRYBA4
|
crystallin beta A4 |
| chr11_-_31804067 | 0.50 |
ENST00000639548.1
ENST00000640125.1 ENST00000481563.6 ENST00000639079.1 ENST00000638762.1 ENST00000638346.1 |
PAX6
|
paired box 6 |
| chr5_+_148268741 | 0.49 |
ENST00000398450.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chrX_+_103330206 | 0.49 |
ENST00000372666.1
ENST00000332431.5 |
TCEAL7
|
transcription elongation factor A like 7 |
| chr17_-_15619512 | 0.49 |
ENST00000395667.7
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr22_+_42553862 | 0.49 |
ENST00000340239.8
ENST00000327678.10 |
SERHL2
|
serine hydrolase like 2 |
| chr1_+_54806063 | 0.49 |
ENST00000358193.7
ENST00000371273.4 |
LEXM
|
lymphocyte expansion molecule |
| chr5_+_148268830 | 0.49 |
ENST00000511106.5
|
SPINK13
|
serine peptidase inhibitor Kazal type 13 |
| chr3_+_111998915 | 0.48 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr12_-_104050112 | 0.48 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr3_-_24495532 | 0.47 |
ENST00000643772.1
ENST00000642307.1 ENST00000645139.1 |
THRB
|
thyroid hormone receptor beta |
| chr12_-_110502065 | 0.47 |
ENST00000447578.6
ENST00000546588.1 ENST00000360579.11 ENST00000549578.6 ENST00000549970.5 |
VPS29
|
VPS29 retromer complex component |
| chr2_+_233712905 | 0.46 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr14_-_80231052 | 0.46 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr5_-_1801284 | 0.45 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr12_-_70754631 | 0.45 |
ENST00000440835.6
ENST00000549308.5 ENST00000550661.1 ENST00000378778.5 |
PTPRR
|
protein tyrosine phosphatase receptor type R |
| chr13_-_51845169 | 0.44 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr8_-_101206064 | 0.44 |
ENST00000518336.5
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr4_+_85604146 | 0.43 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_-_4994696 | 0.43 |
ENST00000400186.7
ENST00000602723.5 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chrX_-_154805386 | 0.43 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chrX_+_47218232 | 0.43 |
ENST00000457458.6
ENST00000522883.1 |
CDK16
|
cyclin dependent kinase 16 |
| chr4_+_87608529 | 0.43 |
ENST00000651931.1
|
DSPP
|
dentin sialophosphoprotein |
| chr4_+_37960397 | 0.43 |
ENST00000504686.2
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr20_+_841238 | 0.43 |
ENST00000541082.2
|
FAM110A
|
family with sequence similarity 110 member A |
| chr8_-_13276491 | 0.42 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr1_-_119811458 | 0.42 |
ENST00000256585.10
ENST00000354219.5 ENST00000369401.4 |
REG4
|
regenerating family member 4 |
| chr2_-_61017174 | 0.42 |
ENST00000407787.5
ENST00000398658.2 |
PUS10
|
pseudouridine synthase 10 |
| chr12_-_11031407 | 0.42 |
ENST00000390675.2
|
TAS2R31
|
taste 2 receptor member 31 |
| chr7_+_142598016 | 0.42 |
ENST00000620773.1
|
TRBV16
|
T cell receptor beta variable 16 |
| chr3_+_127193131 | 0.41 |
ENST00000624688.2
|
C3orf56
|
chromosome 3 open reading frame 56 |
| chr19_-_55831987 | 0.41 |
ENST00000592953.5
ENST00000589093.5 |
NLRP11
|
NLR family pyrin domain containing 11 |
| chr2_-_88979016 | 0.41 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr15_-_79090760 | 0.40 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr19_-_41688258 | 0.40 |
ENST00000401731.6
ENST00000006724.7 |
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr14_-_106811131 | 0.39 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr22_+_19760714 | 0.39 |
ENST00000649276.2
|
TBX1
|
T-box transcription factor 1 |
| chr6_+_29111560 | 0.38 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr14_+_21868822 | 0.38 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr5_+_141421020 | 0.37 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr8_-_33567118 | 0.36 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr8_-_4994904 | 0.36 |
ENST00000635120.2
|
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr1_-_248645278 | 0.36 |
ENST00000641268.1
|
OR2T35
|
olfactory receptor family 2 subfamily T member 35 |
| chr19_-_633500 | 0.35 |
ENST00000588649.7
|
POLRMT
|
RNA polymerase mitochondrial |
| chr18_+_35041387 | 0.35 |
ENST00000538170.6
ENST00000300249.10 ENST00000588910.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr2_+_216412414 | 0.35 |
ENST00000430374.5
ENST00000357276.9 ENST00000444508.5 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a like 1 |
| chrX_-_48957871 | 0.34 |
ENST00000610466.4
|
OTUD5
|
OTU deubiquitinase 5 |
| chr14_-_70416973 | 0.34 |
ENST00000555276.5
ENST00000617124.4 |
COX16
SYNJ2BP-COX16
|
cytochrome c oxidase assembly factor COX16 SYNJ2BP-COX16 readthrough |
| chr19_+_8052752 | 0.33 |
ENST00000315626.6
ENST00000253451.9 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr11_-_117876719 | 0.33 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr15_-_64989435 | 0.33 |
ENST00000433215.6
ENST00000558415.5 ENST00000557795.5 |
SPG21
|
SPG21 abhydrolase domain containing, maspardin |
| chr1_-_108192818 | 0.33 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 member 24 |
| chr19_+_8053000 | 0.33 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25 |
| chr7_-_135977308 | 0.33 |
ENST00000435723.1
ENST00000393085.4 |
MTPN
|
myotrophin |
| chr13_-_28100556 | 0.32 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr12_-_95996302 | 0.32 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr12_-_10435940 | 0.32 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr2_+_195656734 | 0.31 |
ENST00000409086.7
|
SLC39A10
|
solute carrier family 39 member 10 |
| chrX_+_30243715 | 0.31 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr5_+_141343818 | 0.31 |
ENST00000619750.1
ENST00000253812.8 |
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr17_-_76167078 | 0.31 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr9_-_125484490 | 0.30 |
ENST00000444226.1
|
MAPKAP1
|
MAPK associated protein 1 |
| chr1_-_247458105 | 0.30 |
ENST00000641149.1
ENST00000641527.1 |
OR2B11
|
olfactory receptor family 2 subfamily B member 11 |
| chr11_-_117876892 | 0.30 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_+_3658193 | 0.29 |
ENST00000252505.4
|
ALLC
|
allantoicase |
| chr16_+_24539536 | 0.29 |
ENST00000568015.5
ENST00000319715.10 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr12_-_55927756 | 0.28 |
ENST00000549939.1
|
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr22_+_37282464 | 0.28 |
ENST00000402997.5
ENST00000405206.3 ENST00000248901.11 |
CYTH4
|
cytohesin 4 |
| chr3_+_68006224 | 0.28 |
ENST00000496687.1
|
TAFA1
|
TAFA chemokine like family member 1 |
| chr11_-_105023136 | 0.28 |
ENST00000526056.5
ENST00000531367.5 ENST00000456094.1 ENST00000444749.6 ENST00000393141.6 ENST00000418434.5 ENST00000260315.8 |
CASP5
|
caspase 5 |
| chr1_+_160081529 | 0.28 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr12_-_69699455 | 0.27 |
ENST00000266661.8
|
BEST3
|
bestrophin 3 |
| chr7_-_36724380 | 0.27 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.7 | 5.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.7 | 7.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.4 | 1.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 0.8 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.3 | 1.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 1.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.2 | 6.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.0 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.2 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 1.7 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 3.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.2 | 2.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.2 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.7 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 1.0 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 1.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.9 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 1.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.4 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 0.4 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 2.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 1.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.8 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.1 | 0.2 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.2 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 0.5 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 6.0 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 2.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 1.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.5 | GO:0060717 | chorion development(GO:0060717) |
| 0.0 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 1.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.8 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 1.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 1.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 4.7 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.9 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 3.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.7 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 5.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.8 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.5 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 1.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.2 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 3.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 7.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 4.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 4.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.8 | 5.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 1.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 2.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.3 | 1.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 1.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 0.7 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.2 | 0.7 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 4.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.5 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.2 | 6.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 5.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 1.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 6.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 3.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 5.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 6.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |