Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for SOX30
Z-value: 0.35
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.11 | SOX30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg38_v1_chr5_-_157652364_157652385, hg38_v1_chr5_-_157652432_157652443 | -0.20 | 2.9e-01 | Click! |
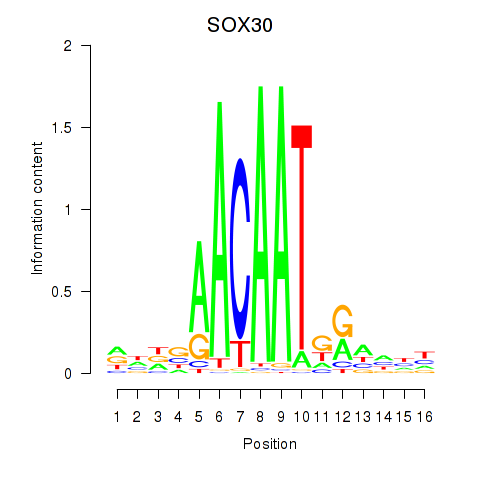
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_27981805 | 1.46 |
ENST00000673512.1
ENST00000672877.1 ENST00000480504.1 |
ODAD2
|
outer dynein arm docking complex subunit 2 |
| chr6_+_131637296 | 0.98 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr13_-_85799400 | 0.86 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr15_-_48963912 | 0.80 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr12_-_49187369 | 0.77 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr6_-_139291987 | 0.76 |
ENST00000358430.8
|
TXLNB
|
taxilin beta |
| chr12_-_10130241 | 0.74 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr1_+_47023659 | 0.64 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr3_+_181711915 | 0.52 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr4_+_76435216 | 0.50 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3 |
| chr12_-_10130143 | 0.47 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr3_-_149221811 | 0.43 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr11_+_63369779 | 0.42 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr5_-_147081428 | 0.41 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr5_-_160852200 | 0.41 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr5_+_140855882 | 0.40 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr8_+_24294044 | 0.39 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr11_-_85719160 | 0.38 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chr12_-_10130082 | 0.37 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr8_+_104339796 | 0.34 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr10_+_68721012 | 0.33 |
ENST00000536391.5
ENST00000630771.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr5_-_147081462 | 0.31 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr11_+_36296281 | 0.31 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr11_-_85719111 | 0.30 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr6_-_134318097 | 0.28 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_26152188 | 0.27 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr7_+_102285125 | 0.27 |
ENST00000536178.3
|
SH2B2
|
SH2B adaptor protein 2 |
| chr9_-_71768386 | 0.25 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr1_+_244835616 | 0.24 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr11_-_85719045 | 0.23 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr20_-_51768327 | 0.22 |
ENST00000311637.9
ENST00000338821.6 |
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr8_+_24294107 | 0.21 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr6_+_125919210 | 0.21 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_125919296 | 0.20 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_-_11643871 | 0.20 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chrX_+_70444827 | 0.20 |
ENST00000374360.8
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr2_+_231708511 | 0.19 |
ENST00000341369.11
ENST00000409115.8 ENST00000409683.5 |
PTMA
|
prothymosin alpha |
| chrX_-_117973579 | 0.18 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr20_+_38962299 | 0.15 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr1_-_156859087 | 0.15 |
ENST00000368195.4
|
INSRR
|
insulin receptor related receptor |
| chr5_+_141408000 | 0.15 |
ENST00000616430.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr15_-_52652031 | 0.15 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr3_-_165837412 | 0.14 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr7_-_93226449 | 0.13 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr2_+_222671651 | 0.13 |
ENST00000446656.4
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr2_-_37671633 | 0.13 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr13_-_41019289 | 0.12 |
ENST00000239882.7
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr2_-_157325808 | 0.12 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr2_-_37672178 | 0.11 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr11_-_102452758 | 0.11 |
ENST00000398136.7
ENST00000361236.7 |
TMEM123
|
transmembrane protein 123 |
| chr17_+_69502397 | 0.11 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_-_37672448 | 0.11 |
ENST00000611976.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr3_-_18424533 | 0.11 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr4_+_186227501 | 0.10 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr5_+_141245384 | 0.10 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr1_-_167914089 | 0.10 |
ENST00000476818.2
ENST00000367848.1 ENST00000367851.9 ENST00000545172.5 |
ADCY10
|
adenylate cyclase 10 |
| chr3_-_71305986 | 0.09 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_8644782 | 0.08 |
ENST00000544814.6
|
SSUH2
|
ssu-2 homolog |
| chrX_-_24672654 | 0.07 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_144070313 | 0.07 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr1_+_240091866 | 0.07 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr1_+_180928133 | 0.07 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr1_+_185045515 | 0.07 |
ENST00000367509.8
ENST00000367510.8 |
RNF2
|
ring finger protein 2 |
| chr19_-_43935234 | 0.07 |
ENST00000269973.10
|
ZNF45
|
zinc finger protein 45 |
| chr16_+_68539213 | 0.07 |
ENST00000570495.5
ENST00000564323.5 ENST00000562156.5 ENST00000573685.5 ENST00000563169.7 ENST00000611381.4 |
ZFP90
|
ZFP90 zinc finger protein |
| chr1_+_185734362 | 0.07 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr14_-_89954659 | 0.06 |
ENST00000555070.1
ENST00000316738.12 ENST00000538485.6 ENST00000556609.5 |
ENSG00000259053.1
EFCAB11
|
novel transcript EF-hand calcium binding domain 11 |
| chr4_+_70592253 | 0.06 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr15_+_56918612 | 0.06 |
ENST00000438423.6
ENST00000267811.9 ENST00000333725.10 ENST00000559609.5 |
TCF12
|
transcription factor 12 |
| chr11_-_18526885 | 0.06 |
ENST00000251968.4
ENST00000536719.5 |
TSG101
|
tumor susceptibility 101 |
| chr2_-_162074182 | 0.05 |
ENST00000360534.8
|
DPP4
|
dipeptidyl peptidase 4 |
| chr5_+_141408032 | 0.05 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr2_-_162074394 | 0.05 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4 |
| chr2_+_178320228 | 0.05 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr14_-_89954518 | 0.05 |
ENST00000556005.1
ENST00000555872.5 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr11_+_196761 | 0.05 |
ENST00000325113.9
|
ODF3
|
outer dense fiber of sperm tails 3 |
| chr1_+_40477248 | 0.05 |
ENST00000372706.6
|
ZFP69
|
ZFP69 zinc finger protein |
| chr5_+_141172637 | 0.04 |
ENST00000231137.6
|
PCDHB7
|
protocadherin beta 7 |
| chr5_+_141213919 | 0.04 |
ENST00000341948.6
|
PCDHB13
|
protocadherin beta 13 |
| chr4_+_70592295 | 0.04 |
ENST00000449493.2
|
AMBN
|
ameloblastin |
| chr14_+_35826298 | 0.04 |
ENST00000216807.12
|
BRMS1L
|
BRMS1 like transcriptional repressor |
| chr11_-_102705737 | 0.03 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr5_-_135399211 | 0.03 |
ENST00000312469.8
ENST00000423969.6 |
MACROH2A1
|
macroH2A.1 histone |
| chr22_-_43089375 | 0.03 |
ENST00000266254.12
|
TTLL1
|
tubulin tyrosine ligase like 1 |
| chr4_-_175812746 | 0.03 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chrX_-_143636094 | 0.03 |
ENST00000356928.2
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr15_+_56918763 | 0.03 |
ENST00000557843.5
|
TCF12
|
transcription factor 12 |
| chr5_+_141392616 | 0.03 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr4_-_68245683 | 0.02 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr7_-_149028452 | 0.01 |
ENST00000413966.1
ENST00000652332.1 |
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr7_-_83162899 | 0.01 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chrX_+_71283577 | 0.01 |
ENST00000420903.6
ENST00000373856.8 ENST00000678437.1 ENST00000678830.1 ENST00000677879.1 ENST00000373841.5 ENST00000276079.13 ENST00000676797.1 ENST00000678660.1 ENST00000678231.1 ENST00000677612.1 ENST00000413858.5 ENST00000450092.6 |
NONO
|
non-POU domain containing octamer binding |
| chr1_-_21176836 | 0.01 |
ENST00000634879.2
ENST00000400422.6 ENST00000602326.5 ENST00000411888.5 ENST00000438975.5 ENST00000374935.7 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr12_-_10098940 | 0.01 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr17_-_10469558 | 0.01 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr7_+_144000320 | 0.01 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr3_+_141262614 | 0.01 |
ENST00000504264.5
|
PXYLP1
|
2-phosphoxylose phosphatase 1 |
| chr11_+_65890627 | 0.01 |
ENST00000312579.4
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr1_-_23369813 | 0.01 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436 |
| chr2_+_206939515 | 0.00 |
ENST00000272852.4
|
CPO
|
carboxypeptidase O |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 1.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.5 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0036343 | negative regulation of extracellular matrix disassembly(GO:0010716) regulation of cell-cell adhesion mediated by integrin(GO:0033632) psychomotor behavior(GO:0036343) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |