Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
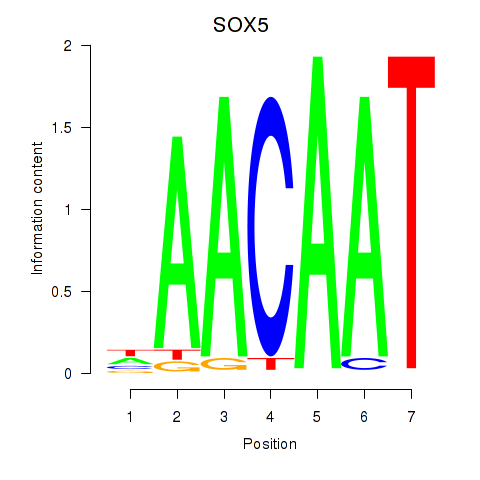
Results for SOX5
Z-value: 1.22
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.19 | SOX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg38_v1_chr12_-_23951020_23951037 | 0.12 | 5.2e-01 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_55376818 | 6.29 |
ENST00000291934.4
|
TMEM190
|
transmembrane protein 190 |
| chr4_+_164754045 | 6.23 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr6_-_32589833 | 5.91 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr7_-_16881967 | 5.69 |
ENST00000402239.7
ENST00000310398.7 ENST00000414935.1 |
AGR3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr4_+_164754116 | 5.42 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr15_+_70936487 | 4.98 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr3_-_19946970 | 4.68 |
ENST00000344838.8
|
EFHB
|
EF-hand domain family member B |
| chr3_+_181711915 | 4.48 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr20_-_3781440 | 4.40 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr7_-_131556602 | 4.35 |
ENST00000322985.9
ENST00000378555.8 |
PODXL
|
podocalyxin like |
| chr15_-_48963912 | 3.82 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr7_-_117873420 | 3.81 |
ENST00000160373.8
|
CTTNBP2
|
cortactin binding protein 2 |
| chr1_-_36440873 | 3.63 |
ENST00000433045.6
|
OSCP1
|
organic solute carrier partner 1 |
| chr7_+_114414809 | 3.45 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr13_+_50015254 | 3.42 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr13_+_50015438 | 3.42 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr1_-_39901996 | 3.25 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chrX_-_117973579 | 3.24 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr4_+_25655822 | 3.21 |
ENST00000504570.5
ENST00000382051.8 |
SLC34A2
|
solute carrier family 34 member 2 |
| chr11_+_36375978 | 2.93 |
ENST00000378867.7
|
PRR5L
|
proline rich 5 like |
| chrY_+_12904102 | 2.92 |
ENST00000360160.8
ENST00000454054.5 |
DDX3Y
|
DEAD-box helicase 3 Y-linked |
| chr17_+_38705482 | 2.86 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chr14_-_91947654 | 2.52 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr3_+_41194741 | 2.46 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr11_+_36296281 | 2.39 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr14_-_91947383 | 2.37 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr12_-_110920568 | 2.32 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr10_+_97319250 | 2.31 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1 |
| chr2_-_156342348 | 2.31 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr10_-_97334698 | 2.27 |
ENST00000371019.4
|
FRAT2
|
FRAT regulator of WNT signaling pathway 2 |
| chr1_+_61081728 | 2.22 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr12_-_85836372 | 2.19 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr1_-_217076889 | 2.13 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr14_-_89417148 | 2.13 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chr9_-_127950716 | 2.12 |
ENST00000373084.8
|
FAM102A
|
family with sequence similarity 102 member A |
| chr10_-_114684612 | 2.04 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_-_39901861 | 2.03 |
ENST00000372816.3
ENST00000372815.1 |
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr1_+_87328860 | 2.00 |
ENST00000370544.10
|
LMO4
|
LIM domain only 4 |
| chrX_-_117973717 | 1.99 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr15_-_40108861 | 1.98 |
ENST00000354670.9
ENST00000559701.5 ENST00000557870.1 ENST00000558774.5 |
BMF
|
Bcl2 modifying factor |
| chr9_-_122227525 | 1.77 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr10_-_32378720 | 1.77 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr4_-_104494882 | 1.74 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4 |
| chr2_+_69013170 | 1.71 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr16_+_2033264 | 1.71 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr4_+_41612892 | 1.59 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_-_16356538 | 1.54 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr18_-_55510753 | 1.52 |
ENST00000543082.5
|
TCF4
|
transcription factor 4 |
| chr20_+_33562365 | 1.49 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr2_+_69013379 | 1.47 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr3_-_52452828 | 1.46 |
ENST00000496590.1
|
TNNC1
|
troponin C1, slow skeletal and cardiac type |
| chr8_-_71361860 | 1.44 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chrX_-_112840815 | 1.43 |
ENST00000304758.5
ENST00000371959.9 |
AMOT
|
angiomotin |
| chr7_-_47582076 | 1.42 |
ENST00000311160.14
|
TNS3
|
tensin 3 |
| chr1_-_85578345 | 1.41 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr6_-_79234619 | 1.39 |
ENST00000344726.9
ENST00000275036.11 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr18_+_58341038 | 1.38 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr8_-_107497909 | 1.36 |
ENST00000517746.6
|
ANGPT1
|
angiopoietin 1 |
| chr6_-_79234713 | 1.35 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr2_+_231708511 | 1.33 |
ENST00000341369.11
ENST00000409115.8 ENST00000409683.5 |
PTMA
|
prothymosin alpha |
| chr12_+_20815672 | 1.32 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr3_+_14402592 | 1.32 |
ENST00000622186.5
ENST00000621751.4 |
SLC6A6
|
solute carrier family 6 member 6 |
| chr18_-_55321986 | 1.30 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr8_-_71362054 | 1.29 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_-_127980976 | 1.27 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr10_+_113125536 | 1.22 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr6_+_125919210 | 1.22 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr9_-_15510991 | 1.20 |
ENST00000380715.5
ENST00000380716.8 ENST00000380738.8 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr9_-_37465402 | 1.18 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr6_+_122399536 | 1.18 |
ENST00000452194.5
|
HSF2
|
heat shock transcription factor 2 |
| chr7_+_114414997 | 1.17 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr9_-_15510954 | 1.16 |
ENST00000380733.9
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr6_+_125919296 | 1.16 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr8_-_40897814 | 1.15 |
ENST00000297737.11
ENST00000315769.11 |
ZMAT4
|
zinc finger matrin-type 4 |
| chr14_-_91946989 | 1.15 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr20_-_21397513 | 1.15 |
ENST00000351817.5
|
NKX2-4
|
NK2 homeobox 4 |
| chr6_+_122399621 | 1.14 |
ENST00000368455.9
|
HSF2
|
heat shock transcription factor 2 |
| chr10_+_35126923 | 1.11 |
ENST00000374726.7
|
CREM
|
cAMP responsive element modulator |
| chr18_-_55321640 | 1.09 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr10_-_60389833 | 1.09 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr2_-_159616442 | 1.09 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr10_-_60572599 | 1.09 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr3_-_115071333 | 1.08 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_187448244 | 1.06 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr1_+_147541491 | 1.06 |
ENST00000683836.1
ENST00000234739.8 |
BCL9
|
BCL9 transcription coactivator |
| chr9_-_120477354 | 1.04 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr11_+_111937320 | 1.04 |
ENST00000440460.7
|
DIXDC1
|
DIX domain containing 1 |
| chr9_-_123268538 | 1.01 |
ENST00000360998.3
ENST00000348403.10 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr1_+_33256479 | 1.00 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr3_+_171843337 | 0.99 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr5_+_173889337 | 0.97 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr6_-_139291987 | 0.97 |
ENST00000358430.8
|
TXLNB
|
taxilin beta |
| chr3_-_11643871 | 0.95 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr3_-_65597886 | 0.95 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr18_-_55322215 | 0.92 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr10_+_68721012 | 0.92 |
ENST00000536391.5
ENST00000630771.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr15_+_83447328 | 0.89 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr2_+_69013414 | 0.89 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr15_-_37099306 | 0.89 |
ENST00000557796.6
ENST00000397620.6 |
MEIS2
|
Meis homeobox 2 |
| chr10_+_102776237 | 0.88 |
ENST00000369889.5
|
WBP1L
|
WW domain binding protein 1 like |
| chr3_+_189789672 | 0.87 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr15_+_83447411 | 0.86 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr9_-_71768386 | 0.86 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr21_-_17819343 | 0.85 |
ENST00000284881.9
|
C21orf91
|
chromosome 21 open reading frame 91 |
| chr9_-_14314132 | 0.85 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chr6_+_43059857 | 0.82 |
ENST00000259708.7
ENST00000472792.1 ENST00000479388.5 ENST00000460283.1 ENST00000394056.6 |
KLC4
|
kinesin light chain 4 |
| chr15_+_43792839 | 0.81 |
ENST00000409614.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr5_-_38595396 | 0.80 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha |
| chr2_+_86441341 | 0.76 |
ENST00000312912.10
ENST00000409064.5 |
KDM3A
|
lysine demethylase 3A |
| chr13_+_98143410 | 0.75 |
ENST00000596580.2
ENST00000376581.9 |
FARP1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr3_-_18424533 | 0.75 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr20_-_18497218 | 0.75 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr3_-_114624193 | 0.75 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_+_68721207 | 0.74 |
ENST00000538031.5
ENST00000543719.5 ENST00000539539.5 ENST00000265872.11 ENST00000543225.5 ENST00000536012.5 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr4_-_2262082 | 0.74 |
ENST00000337190.7
|
MXD4
|
MAX dimerization protein 4 |
| chr10_-_14572123 | 0.74 |
ENST00000378465.7
ENST00000452706.6 ENST00000622567.4 ENST00000378458.6 |
FAM107B
|
family with sequence similarity 107 member B |
| chr4_+_169660062 | 0.73 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr17_+_38705243 | 0.72 |
ENST00000621332.5
|
MLLT6
|
MLLT6, PHD finger containing |
| chrX_-_13817027 | 0.71 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr21_-_17819386 | 0.71 |
ENST00000400559.7
ENST00000400558.7 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr7_+_117480011 | 0.70 |
ENST00000649406.1
ENST00000648260.1 ENST00000003084.11 |
CFTR
|
CF transmembrane conductance regulator |
| chr12_-_122500924 | 0.70 |
ENST00000633063.3
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr15_+_98648502 | 0.67 |
ENST00000650285.1
ENST00000649865.1 |
IGF1R
|
insulin like growth factor 1 receptor |
| chr12_-_122500947 | 0.67 |
ENST00000672018.1
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr12_-_389249 | 0.65 |
ENST00000535014.5
ENST00000543507.1 ENST00000544760.1 ENST00000399788.7 |
KDM5A
|
lysine demethylase 5A |
| chr3_+_113211459 | 0.65 |
ENST00000495514.5
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr6_+_118894144 | 0.65 |
ENST00000229595.6
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr3_-_71306012 | 0.64 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr1_+_43172324 | 0.64 |
ENST00000528956.5
ENST00000610710.4 ENST00000372492.9 ENST00000529956.5 |
CFAP57
|
cilia and flagella associated protein 57 |
| chr1_+_164559739 | 0.63 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr20_+_6007245 | 0.63 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr15_-_85794902 | 0.63 |
ENST00000337975.6
|
KLHL25
|
kelch like family member 25 |
| chr8_-_69833338 | 0.62 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr6_-_116060859 | 0.62 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr14_+_23321543 | 0.62 |
ENST00000556821.5
|
PABPN1
|
poly(A) binding protein nuclear 1 |
| chr12_+_389334 | 0.62 |
ENST00000540180.5
ENST00000422000.5 ENST00000535052.5 |
CCDC77
|
coiled-coil domain containing 77 |
| chr15_-_45201094 | 0.61 |
ENST00000561278.1
ENST00000290894.12 |
SHF
|
Src homology 2 domain containing F |
| chr10_+_68106109 | 0.61 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr1_+_167329044 | 0.60 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr9_+_68780029 | 0.60 |
ENST00000394264.7
|
PABIR1
|
PP2A Aalpha (PPP2R1A) and B55A (PPP2R2A) interacting phosphatase regulator 1 |
| chr19_+_37907200 | 0.60 |
ENST00000222345.11
|
SIPA1L3
|
signal induced proliferation associated 1 like 3 |
| chr2_+_86720282 | 0.60 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr5_+_68292562 | 0.59 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr6_+_156777882 | 0.58 |
ENST00000350026.10
ENST00000647938.1 ENST00000674298.1 |
ARID1B
|
AT-rich interaction domain 1B |
| chr5_+_140855882 | 0.58 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr1_+_26111139 | 0.57 |
ENST00000619836.4
ENST00000444713.5 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr9_+_2159672 | 0.56 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_-_143635081 | 0.56 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr9_+_27109393 | 0.56 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr4_+_94995919 | 0.55 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr16_+_31074390 | 0.54 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr4_+_41612702 | 0.54 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_-_14314567 | 0.54 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr1_+_206834347 | 0.54 |
ENST00000340758.7
|
IL19
|
interleukin 19 |
| chr5_+_139308095 | 0.53 |
ENST00000515833.2
|
MATR3
|
matrin 3 |
| chr20_-_47356721 | 0.53 |
ENST00000262975.8
ENST00000446994.6 ENST00000355972.8 ENST00000396281.8 ENST00000619049.4 ENST00000611941.4 ENST00000372023.7 |
ZMYND8
|
zinc finger MYND-type containing 8 |
| chrX_+_70444827 | 0.53 |
ENST00000374360.8
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr1_+_164559766 | 0.52 |
ENST00000367897.5
ENST00000559240.5 |
PBX1
|
PBX homeobox 1 |
| chr6_+_43059614 | 0.52 |
ENST00000453940.6
ENST00000347162.10 ENST00000479632.5 ENST00000470728.5 ENST00000458460.6 |
KLC4
|
kinesin light chain 4 |
| chr3_-_71305986 | 0.52 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr12_-_122500520 | 0.51 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr9_-_95509241 | 0.51 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chr15_+_66702219 | 0.51 |
ENST00000288840.10
|
SMAD6
|
SMAD family member 6 |
| chr15_-_40874216 | 0.51 |
ENST00000220507.5
|
RHOV
|
ras homolog family member V |
| chr1_-_182391363 | 0.51 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr1_-_161038907 | 0.50 |
ENST00000318289.14
ENST00000368023.7 ENST00000423014.3 ENST00000368024.5 |
TSTD1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr3_+_113211539 | 0.49 |
ENST00000682979.1
ENST00000485230.5 |
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr9_-_120714457 | 0.49 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chr4_+_159267737 | 0.49 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr6_+_156777366 | 0.47 |
ENST00000636930.2
|
ARID1B
|
AT-rich interaction domain 1B |
| chr1_-_52552994 | 0.47 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr11_-_7020235 | 0.47 |
ENST00000536068.5
ENST00000278314.5 |
ZNF214
|
zinc finger protein 214 |
| chr2_+_119759875 | 0.47 |
ENST00000263708.7
|
PTPN4
|
protein tyrosine phosphatase non-receptor type 4 |
| chr19_+_7516081 | 0.46 |
ENST00000597229.2
|
ZNF358
|
zinc finger protein 358 |
| chr4_-_185810894 | 0.46 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_86360010 | 0.45 |
ENST00000641911.1
ENST00000641072.1 ENST00000359221.8 ENST00000640490.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr15_-_52652031 | 0.45 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr1_+_177170916 | 0.45 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_-_153670966 | 0.45 |
ENST00000368681.1
ENST00000361891.9 ENST00000615950.4 |
ILF2
|
interleukin enhancer binding factor 2 |
| chr8_+_56211686 | 0.45 |
ENST00000521831.5
ENST00000303759.3 ENST00000517636.5 ENST00000517933.5 ENST00000355315.8 ENST00000518801.5 ENST00000523975.5 ENST00000396723.9 ENST00000523061.5 ENST00000521524.5 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr12_+_11649666 | 0.44 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr12_-_51083582 | 0.44 |
ENST00000548206.1
ENST00000546935.5 ENST00000228515.6 ENST00000548981.5 |
CSRNP2
|
cysteine and serine rich nuclear protein 2 |
| chr9_-_14314519 | 0.44 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr10_+_21524627 | 0.43 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr7_-_7535863 | 0.42 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr2_+_109898685 | 0.42 |
ENST00000480744.2
|
LIMS3
|
LIM zinc finger domain containing 3 |
| chr5_-_147081428 | 0.40 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr3_+_173398438 | 0.40 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr9_-_137302264 | 0.39 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr5_+_139341875 | 0.39 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_226063466 | 0.39 |
ENST00000666609.1
ENST00000661429.1 |
H3-3A
|
H3.3 histone A |
| chr1_-_50423431 | 0.39 |
ENST00000404795.4
|
DMRTA2
|
DMRT like family A2 |
| chr2_-_70248598 | 0.38 |
ENST00000445587.5
ENST00000433529.7 ENST00000415783.6 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
| chr6_-_90296908 | 0.38 |
ENST00000537989.5
|
BACH2
|
BTB domain and CNC homolog 2 |
| chr8_+_143213192 | 0.38 |
ENST00000622500.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr10_+_35127162 | 0.38 |
ENST00000354759.7
|
CREM
|
cAMP responsive element modulator |
| chr2_-_157325808 | 0.38 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr10_+_48306698 | 0.38 |
ENST00000374179.8
|
MAPK8
|
mitogen-activated protein kinase 8 |
| chr6_+_104957099 | 0.37 |
ENST00000345080.5
|
LIN28B
|
lin-28 homolog B |
| chr5_-_147081462 | 0.37 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 1.5 | 5.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.1 | 4.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.9 | 4.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.8 | 2.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.7 | 5.3 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.5 | 2.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 6.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 6.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.5 | 1.5 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.5 | 1.4 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.4 | 2.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 2.5 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.4 | 3.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 0.9 | GO:1902161 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.3 | 5.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.5 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 2.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 4.5 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.3 | 2.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 0.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.3 | 0.8 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 0.5 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.2 | 0.7 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.9 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.2 | 2.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 1.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.2 | 1.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.7 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 2.0 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 4.6 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.5 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.5 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.5 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 1.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 1.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.5 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.4 | GO:0071503 | positive regulation of lipoprotein particle clearance(GO:0010986) response to heparin(GO:0071503) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 2.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 2.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.6 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 1.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.8 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.1 | 4.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 1.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 1.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.6 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 1.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 2.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 2.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 3.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:1901223 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 5.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 5.0 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 4.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.5 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.7 | 6.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 2.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 5.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 4.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 1.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 5.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 2.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.9 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 1.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 2.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 3.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 4.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 5.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 4.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 6.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 5.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 4.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 0.9 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 2.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 2.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 4.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 5.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 5.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 2.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 3.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 4.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.5 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 7.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 6.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 4.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 5.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 1.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 3.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 3.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 6.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 6.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 6.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 6.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 7.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 2.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 4.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 5.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |