Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
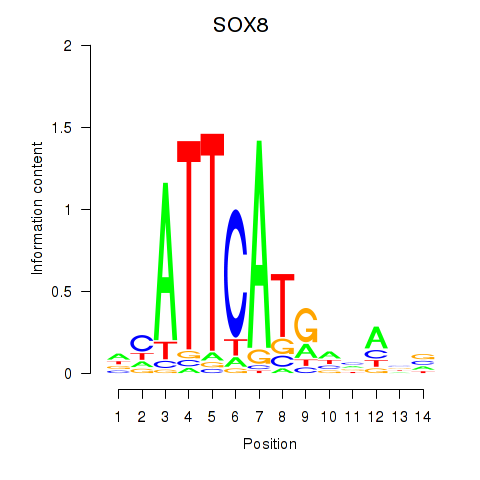
Results for SOX8
Z-value: 1.04
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.10 | SOX8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg38_v1_chr16_+_981762_981782 | -0.06 | 7.7e-01 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_16083695 | 6.43 |
ENST00000510224.5
|
PROM1
|
prominin 1 |
| chr4_-_16083714 | 6.39 |
ENST00000508167.5
|
PROM1
|
prominin 1 |
| chr4_-_16084002 | 5.90 |
ENST00000447510.7
|
PROM1
|
prominin 1 |
| chr12_-_10130241 | 3.63 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr3_+_319683 | 3.39 |
ENST00000620033.4
|
CHL1
|
cell adhesion molecule L1 like |
| chr5_-_160852200 | 3.36 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr6_+_32637419 | 3.31 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr12_-_10130143 | 3.24 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr14_-_106622837 | 3.23 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr6_-_52803807 | 3.19 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr2_+_26401909 | 3.02 |
ENST00000288710.7
|
DRC1
|
dynein regulatory complex subunit 1 |
| chr4_-_99435396 | 2.99 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_71157872 | 2.88 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_10130082 | 2.71 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr4_-_99435336 | 2.59 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_99435134 | 2.55 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_+_50015438 | 2.43 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr16_+_82035245 | 2.15 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr3_-_193554885 | 2.12 |
ENST00000342695.9
|
ATP13A4
|
ATPase 13A4 |
| chr14_-_106185387 | 1.96 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr5_+_141223332 | 1.82 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr3_-_193554952 | 1.77 |
ENST00000392443.7
|
ATP13A4
|
ATPase 13A4 |
| chr13_+_50015254 | 1.73 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr6_+_32637396 | 1.71 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr7_+_48455063 | 1.64 |
ENST00000411975.5
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr8_-_94208548 | 1.61 |
ENST00000027335.8
ENST00000441892.6 ENST00000521491.1 |
CDH17
|
cadherin 17 |
| chr2_-_174846405 | 1.58 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr16_+_57358775 | 1.56 |
ENST00000219235.5
|
CCL22
|
C-C motif chemokine ligand 22 |
| chr2_+_203936755 | 1.52 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr1_+_12746192 | 1.52 |
ENST00000614859.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr5_+_140806929 | 1.37 |
ENST00000378125.4
ENST00000618834.1 ENST00000530339.2 ENST00000512229.6 ENST00000672575.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr14_-_105987068 | 1.30 |
ENST00000390594.3
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr17_+_63998344 | 1.29 |
ENST00000577953.5
ENST00000582540.5 ENST00000579184.5 ENST00000425164.7 ENST00000412177.6 ENST00000583891.5 ENST00000580752.1 |
PRR29
|
proline rich 29 |
| chr11_+_6876625 | 1.20 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr14_+_94612383 | 1.17 |
ENST00000393080.8
ENST00000555820.1 ENST00000393078.5 ENST00000467132.5 |
SERPINA3
|
serpin family A member 3 |
| chr10_+_7703300 | 1.15 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr20_+_59628609 | 1.15 |
ENST00000541461.5
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_+_95051671 | 1.14 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr16_-_2329687 | 1.11 |
ENST00000567910.1
|
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr7_-_120858303 | 1.10 |
ENST00000415871.5
ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr18_-_55585773 | 1.10 |
ENST00000563824.5
ENST00000626425.2 ENST00000566514.5 ENST00000568673.5 ENST00000562847.5 ENST00000568147.5 |
TCF4
|
transcription factor 4 |
| chr15_-_65422894 | 1.10 |
ENST00000352385.3
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4 |
| chr19_-_45178200 | 1.07 |
ENST00000592647.1
ENST00000006275.8 ENST00000588062.5 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr5_-_180072086 | 1.05 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr12_-_14951106 | 1.00 |
ENST00000541644.5
ENST00000545895.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chrX_-_152830721 | 0.99 |
ENST00000370277.5
|
CETN2
|
centrin 2 |
| chr18_-_55586092 | 0.98 |
ENST00000563888.6
ENST00000540999.5 ENST00000627685.2 |
TCF4
|
transcription factor 4 |
| chr17_-_49646581 | 0.95 |
ENST00000510476.5
ENST00000503676.5 |
SPOP
|
speckle type BTB/POZ protein |
| chr1_+_111473972 | 0.95 |
ENST00000369718.4
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr14_-_106025628 | 0.94 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr7_-_93226449 | 0.93 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr4_-_140427635 | 0.92 |
ENST00000325617.10
ENST00000414773.5 |
CLGN
|
calmegin |
| chrX_+_13569593 | 0.91 |
ENST00000361306.6
ENST00000380602.3 |
EGFL6
|
EGF like domain multiple 6 |
| chr8_+_22057857 | 0.88 |
ENST00000517305.4
ENST00000265800.9 ENST00000517418.5 |
DMTN
|
dematin actin binding protein |
| chr1_-_58546693 | 0.88 |
ENST00000456980.5
ENST00000482274.2 ENST00000453710.1 ENST00000371226.8 ENST00000419242.5 ENST00000426139.5 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr14_-_106005574 | 0.84 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr4_-_48080172 | 0.84 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr1_-_48472166 | 0.83 |
ENST00000371847.8
ENST00000396199.7 |
SPATA6
|
spermatogenesis associated 6 |
| chr2_-_32264853 | 0.82 |
ENST00000402280.6
|
NLRC4
|
NLR family CARD domain containing 4 |
| chr1_-_204307391 | 0.82 |
ENST00000637508.1
|
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr15_-_89211803 | 0.81 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr7_-_38249572 | 0.78 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr2_+_169694434 | 0.77 |
ENST00000616481.4
ENST00000616524.4 ENST00000617738.4 ENST00000359744.8 ENST00000438838.5 ENST00000438710.5 ENST00000449906.5 ENST00000498202.6 ENST00000272797.8 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch like family member 23 |
| chr8_-_141367276 | 0.77 |
ENST00000377741.4
|
GPR20
|
G protein-coupled receptor 20 |
| chr9_+_126914760 | 0.77 |
ENST00000424082.6
ENST00000259351.10 ENST00000394022.7 ENST00000394011.7 ENST00000319107.8 |
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr20_-_45547420 | 0.75 |
ENST00000504988.1
|
EPPIN-WFDC6
|
EPPIN-WFDC6 readthrough |
| chr10_-_20897288 | 0.75 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr3_-_49813880 | 0.74 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr3_-_129428590 | 0.74 |
ENST00000503957.1
ENST00000505956.6 ENST00000326085.7 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr6_-_109690500 | 0.74 |
ENST00000448084.6
|
AK9
|
adenylate kinase 9 |
| chr6_-_109690515 | 0.74 |
ENST00000532976.1
|
AK9
|
adenylate kinase 9 |
| chr11_+_124184244 | 0.74 |
ENST00000641546.1
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr4_+_88378842 | 0.73 |
ENST00000264346.12
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr16_+_57735723 | 0.72 |
ENST00000562592.5
ENST00000379661.8 ENST00000566726.5 |
KATNB1
|
katanin regulatory subunit B1 |
| chr16_+_21233672 | 0.72 |
ENST00000311620.7
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr20_+_33562365 | 0.71 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr5_+_95731300 | 0.71 |
ENST00000379982.8
|
RHOBTB3
|
Rho related BTB domain containing 3 |
| chr16_+_53207981 | 0.71 |
ENST00000565803.2
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_91940970 | 0.71 |
ENST00000359028.7
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr1_+_230979064 | 0.68 |
ENST00000366658.6
ENST00000310256.7 ENST00000450711.5 ENST00000435927.5 |
ARV1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr11_-_6619353 | 0.67 |
ENST00000642892.1
ENST00000645620.1 ENST00000533371.6 ENST00000647152.1 ENST00000644810.1 ENST00000299427.12 ENST00000682424.1 ENST00000644218.1 ENST00000528657.2 ENST00000531754.2 |
TPP1
|
tripeptidyl peptidase 1 |
| chr4_+_55346213 | 0.66 |
ENST00000679836.1
ENST00000264228.9 ENST00000679707.1 |
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3 novel protein, SRD5A3-RP11-177J6.1 readthrough |
| chr7_-_32490361 | 0.66 |
ENST00000410044.5
ENST00000450169.7 ENST00000409987.5 ENST00000409782.5 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_-_35880350 | 0.65 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr2_+_232662733 | 0.65 |
ENST00000410095.5
ENST00000611312.1 |
EFHD1
|
EF-hand domain family member D1 |
| chr3_-_142000353 | 0.64 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr20_-_45547373 | 0.62 |
ENST00000354280.9
|
EPPIN
|
epididymal peptidase inhibitor |
| chr19_-_51028015 | 0.61 |
ENST00000319720.11
|
KLK11
|
kallikrein related peptidase 11 |
| chr4_-_86101922 | 0.59 |
ENST00000472236.5
ENST00000641881.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_+_64666156 | 0.59 |
ENST00000344930.7
|
ZNF107
|
zinc finger protein 107 |
| chr11_+_10456186 | 0.59 |
ENST00000528723.5
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr10_+_86654541 | 0.58 |
ENST00000241891.10
ENST00000372071.6 |
OPN4
|
opsin 4 |
| chr6_-_49744434 | 0.58 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr7_+_64794388 | 0.57 |
ENST00000359735.7
|
ZNF138
|
zinc finger protein 138 |
| chr14_+_22112280 | 0.57 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr1_+_111473792 | 0.57 |
ENST00000343534.9
|
C1orf162
|
chromosome 1 open reading frame 162 |
| chr17_-_45262084 | 0.56 |
ENST00000331780.5
|
SPATA32
|
spermatogenesis associated 32 |
| chr3_+_38305936 | 0.55 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22 member 14 |
| chr16_+_67570741 | 0.55 |
ENST00000644753.1
ENST00000642819.1 ENST00000645306.1 |
CTCF
|
CCCTC-binding factor |
| chr3_-_185552554 | 0.54 |
ENST00000424591.6
ENST00000296252.9 |
LIPH
|
lipase H |
| chr16_-_18332662 | 0.54 |
ENST00000541810.5
|
NPIPA8
|
nuclear pore complex interacting protein family member A8 |
| chr9_-_123184233 | 0.54 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr19_+_23117018 | 0.52 |
ENST00000597761.7
|
ZNF730
|
zinc finger protein 730 |
| chr17_+_42980547 | 0.52 |
ENST00000361677.5
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr6_-_49744378 | 0.52 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr19_+_19033575 | 0.50 |
ENST00000392335.6
ENST00000537263.5 ENST00000540707.5 ENST00000535612.6 ENST00000541725.5 ENST00000269932.10 ENST00000546344.5 ENST00000540792.5 ENST00000536098.5 ENST00000541898.5 |
ARMC6
|
armadillo repeat containing 6 |
| chr17_-_40565459 | 0.48 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
C-C motif chemokine receptor 7 |
| chr1_+_248509536 | 0.48 |
ENST00000641501.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr10_-_1737516 | 0.47 |
ENST00000381312.6
|
ADARB2
|
adenosine deaminase RNA specific B2 (inactive) |
| chr1_+_110034607 | 0.47 |
ENST00000369795.8
|
STRIP1
|
striatin interacting protein 1 |
| chr2_-_158380960 | 0.47 |
ENST00000409187.5
|
CCDC148
|
coiled-coil domain containing 148 |
| chr11_-_6006946 | 0.46 |
ENST00000641156.1
ENST00000641835.1 |
OR56A4
|
olfactory receptor family 56 subfamily A member 4 |
| chr17_+_2337622 | 0.46 |
ENST00000574563.5
|
SGSM2
|
small G protein signaling modulator 2 |
| chr3_-_186570308 | 0.45 |
ENST00000446782.5
|
TBCCD1
|
TBCC domain containing 1 |
| chr14_+_22105305 | 0.44 |
ENST00000390453.1
|
TRAV24
|
T cell receptor alpha variable 24 |
| chr3_+_142623386 | 0.44 |
ENST00000337777.7
ENST00000497199.5 |
PLS1
|
plastin 1 |
| chr2_+_102473219 | 0.44 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr12_-_9999176 | 0.43 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr14_-_106875069 | 0.43 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr9_+_2157647 | 0.43 |
ENST00000452193.5
ENST00000324954.10 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_159000174 | 0.43 |
ENST00000367069.7
|
RSPH3
|
radial spoke head 3 |
| chr12_-_9607903 | 0.42 |
ENST00000229402.4
|
KLRB1
|
killer cell lectin like receptor B1 |
| chr15_+_43593054 | 0.41 |
ENST00000453782.5
ENST00000300283.10 ENST00000437924.5 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_-_34929574 | 0.41 |
ENST00000373347.6
|
DLGAP3
|
DLG associated protein 3 |
| chr3_-_54928044 | 0.40 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1 |
| chr12_-_49707220 | 0.40 |
ENST00000550488.5
|
FMNL3
|
formin like 3 |
| chr10_-_6062290 | 0.39 |
ENST00000256876.10
ENST00000379954.5 |
IL2RA
|
interleukin 2 receptor subunit alpha |
| chr5_+_119452467 | 0.38 |
ENST00000509514.6
ENST00000682996.1 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr16_-_70685791 | 0.38 |
ENST00000616026.4
|
MTSS2
|
MTSS I-BAR domain containing 2 |
| chrX_-_19965142 | 0.38 |
ENST00000340625.3
|
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr16_-_66552460 | 0.37 |
ENST00000678015.1
ENST00000569718.6 ENST00000678314.1 ENST00000562484.2 |
TK2
|
thymidine kinase 2 |
| chr1_-_13285154 | 0.37 |
ENST00000357367.6
ENST00000614831.1 |
PRAMEF8
|
PRAME family member 8 |
| chr2_-_169694367 | 0.36 |
ENST00000447353.6
|
CCDC173
|
coiled-coil domain containing 173 |
| chr6_-_31721679 | 0.36 |
ENST00000495859.1
ENST00000375819.3 |
LY6G6C
|
lymphocyte antigen 6 family member G6C |
| chr1_-_244451896 | 0.36 |
ENST00000366535.4
|
ADSS2
|
adenylosuccinate synthase 2 |
| chr17_+_7484357 | 0.36 |
ENST00000674977.2
|
POLR2A
|
RNA polymerase II subunit A |
| chr8_+_106726012 | 0.36 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr1_-_111563934 | 0.36 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr14_+_51860632 | 0.35 |
ENST00000555472.5
ENST00000556766.5 |
GNG2
|
G protein subunit gamma 2 |
| chr13_-_48413105 | 0.35 |
ENST00000620633.5
|
LPAR6
|
lysophosphatidic acid receptor 6 |
| chr6_-_75363003 | 0.35 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr9_-_127771335 | 0.34 |
ENST00000373276.7
ENST00000373277.8 |
SH2D3C
|
SH2 domain containing 3C |
| chr15_-_72271244 | 0.34 |
ENST00000287196.13
|
PARP6
|
poly(ADP-ribose) polymerase family member 6 |
| chrX_-_117973579 | 0.34 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr6_-_149896059 | 0.34 |
ENST00000532335.5
|
RAET1E
|
retinoic acid early transcript 1E |
| chr7_-_83649097 | 0.34 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr9_-_14322320 | 0.33 |
ENST00000606230.2
|
NFIB
|
nuclear factor I B |
| chr1_+_161505438 | 0.33 |
ENST00000271450.12
|
FCGR2A
|
Fc fragment of IgG receptor IIa |
| chr19_-_37207074 | 0.32 |
ENST00000588873.1
|
ENSG00000267360.6
|
novel protein |
| chr4_-_144019287 | 0.32 |
ENST00000638448.1
ENST00000513128.5 ENST00000506516.6 ENST00000429670.3 ENST00000502664.6 |
GYPB
|
glycophorin B (MNS blood group) |
| chr15_-_72118114 | 0.32 |
ENST00000356056.10
ENST00000569314.1 |
MYO9A
|
myosin IXA |
| chr2_+_200440649 | 0.32 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chrX_-_117973717 | 0.31 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr1_+_203305510 | 0.31 |
ENST00000290551.5
|
BTG2
|
BTG anti-proliferation factor 2 |
| chr10_-_80172839 | 0.31 |
ENST00000265447.8
|
ANXA11
|
annexin A11 |
| chr10_+_38094327 | 0.31 |
ENST00000638053.1
ENST00000351773.7 ENST00000361085.9 |
ZNF37A
|
zinc finger protein 37A |
| chr12_+_78036248 | 0.31 |
ENST00000644176.1
|
NAV3
|
neuron navigator 3 |
| chr15_+_62561361 | 0.30 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr17_-_7242156 | 0.30 |
ENST00000571129.5
ENST00000571253.1 ENST00000573928.1 ENST00000302386.10 |
GABARAP
|
GABA type A receptor-associated protein |
| chrY_+_5000226 | 0.30 |
ENST00000333703.8
ENST00000622698.4 ENST00000621505.1 |
PCDH11Y
|
protocadherin 11 Y-linked |
| chr10_-_50279715 | 0.30 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr1_-_111563956 | 0.29 |
ENST00000369717.8
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr17_-_28903017 | 0.29 |
ENST00000394901.7
ENST00000378895.9 |
DHRS13
|
dehydrogenase/reductase 13 |
| chr1_+_15617415 | 0.28 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr11_-_129192198 | 0.28 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr11_-_4339244 | 0.28 |
ENST00000524542.2
|
SSU72P7
|
SSU72 pseudogene 7 |
| chr11_+_7987314 | 0.27 |
ENST00000531572.2
ENST00000651655.1 |
EIF3F
|
eukaryotic translation initiation factor 3 subunit F |
| chr15_+_72118392 | 0.27 |
ENST00000340912.6
|
SENP8
|
SUMO peptidase family member, NEDD8 specific |
| chr9_-_114074969 | 0.27 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr3_-_49422429 | 0.27 |
ENST00000637682.1
ENST00000427987.6 ENST00000636199.1 ENST00000638063.1 ENST00000636865.1 ENST00000458307.6 ENST00000273588.9 ENST00000635808.1 ENST00000636522.1 ENST00000636597.1 ENST00000538581.6 ENST00000395338.7 ENST00000462048.2 |
AMT
|
aminomethyltransferase |
| chr10_-_48274567 | 0.27 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr1_-_31764333 | 0.27 |
ENST00000398542.5
ENST00000373658.8 |
ADGRB2
|
adhesion G protein-coupled receptor B2 |
| chr14_+_55661272 | 0.26 |
ENST00000555573.5
|
KTN1
|
kinectin 1 |
| chr6_-_27132750 | 0.26 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr7_+_80646436 | 0.26 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr4_-_87529359 | 0.26 |
ENST00000458304.2
ENST00000282470.11 |
SPARCL1
|
SPARC like 1 |
| chr3_-_123991352 | 0.26 |
ENST00000184183.8
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr9_-_32573173 | 0.25 |
ENST00000366466.5
|
NDUFB6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr4_+_74308463 | 0.24 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr3_+_156291020 | 0.24 |
ENST00000302490.12
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr10_-_37976589 | 0.24 |
ENST00000302609.8
|
ZNF25
|
zinc finger protein 25 |
| chr8_+_106726115 | 0.24 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr12_-_101830671 | 0.24 |
ENST00000549165.1
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr6_-_75243770 | 0.24 |
ENST00000472311.6
ENST00000460985.1 ENST00000377978.3 ENST00000684430.1 ENST00000509698.6 |
COX7A2
|
cytochrome c oxidase subunit 7A2 |
| chr7_+_144070313 | 0.23 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr15_-_58014097 | 0.23 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr20_-_23826721 | 0.23 |
ENST00000304725.3
|
CST2
|
cystatin SA |
| chr11_+_17260353 | 0.23 |
ENST00000530527.5
|
NUCB2
|
nucleobindin 2 |
| chrX_+_101408198 | 0.23 |
ENST00000316594.6
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr9_-_69014090 | 0.22 |
ENST00000377276.5
|
PRKACG
|
protein kinase cAMP-activated catalytic subunit gamma |
| chr3_+_186570663 | 0.22 |
ENST00000265028.8
|
DNAJB11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr3_+_35642159 | 0.22 |
ENST00000187397.8
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr22_-_36507022 | 0.22 |
ENST00000216187.10
ENST00000397224.9 ENST00000423980.1 |
FOXRED2
|
FAD dependent oxidoreductase domain containing 2 |
| chr11_-_128842467 | 0.21 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr7_+_56064224 | 0.21 |
ENST00000651354.1
ENST00000651586.1 ENST00000652303.1 ENST00000275607.13 ENST00000650735.1 ENST00000395435.7 ENST00000413952.7 ENST00000342190.11 ENST00000437307.6 ENST00000434526.8 ENST00000413756.5 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chr17_-_40782544 | 0.21 |
ENST00000301656.4
|
KRT27
|
keratin 27 |
| chr1_-_12831410 | 0.21 |
ENST00000619922.1
|
PRAMEF11
|
PRAME family member 11 |
| chr1_+_161766309 | 0.21 |
ENST00000679853.1
ENST00000681492.1 ENST00000679886.1 ENST00000367942.4 ENST00000680462.1 ENST00000680633.1 ENST00000681912.1 |
ATF6
|
activating transcription factor 6 |
| chr5_-_157460085 | 0.20 |
ENST00000519499.2
|
ENSG00000285868.1
|
Novel protein |
| chr12_+_55549602 | 0.20 |
ENST00000641569.1
ENST00000641851.1 |
OR6C4
|
olfactory receptor family 6 subfamily C member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.7 | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 2.0 | 8.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 1.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.5 | 1.5 | GO:0061568 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.4 | 9.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.3 | 4.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 1.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 0.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 0.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 0.8 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.7 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.4 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.8 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 2.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.9 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.7 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 3.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 3.4 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.9 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.1 | 3.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 6.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.1 | 0.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.3 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.4 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 1.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.2 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 4.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 2.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 1.0 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.2 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.4 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 0.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 5.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.2 | 0.7 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 5.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.8 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.9 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.7 | 2.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 5.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 1.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 10.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 1.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 18.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.2 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.2 | 0.7 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 0.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.7 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.4 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.5 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 3.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 2.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 5.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 3.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 2.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 1.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 3.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 9.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 5.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |