Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
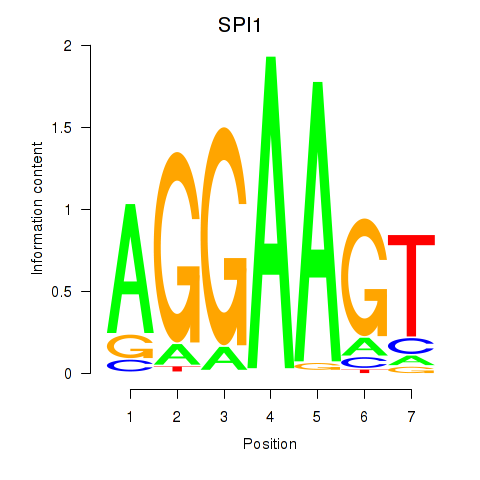
Results for SPI1
Z-value: 1.00
Transcription factors associated with SPI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPI1
|
ENSG00000066336.12 | SPI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPI1 | hg38_v1_chr11_-_47378391_47378410, hg38_v1_chr11_-_47378527_47378555 | 0.36 | 4.7e-02 | Click! |
Activity profile of SPI1 motif
Sorted Z-values of SPI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_63702958 | 3.70 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr3_-_112975018 | 2.70 |
ENST00000471858.5
ENST00000308611.8 ENST00000295863.4 |
CD200R1
|
CD200 receptor 1 |
| chr5_-_180591488 | 2.53 |
ENST00000292641.4
|
SCGB3A1
|
secretoglobin family 3A member 1 |
| chr14_+_96482982 | 2.34 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr17_+_47831608 | 2.28 |
ENST00000269025.9
|
LRRC46
|
leucine rich repeat containing 46 |
| chr3_-_112974912 | 2.20 |
ENST00000440122.6
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr1_-_150765735 | 2.16 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr9_-_114387973 | 1.91 |
ENST00000374088.8
|
AKNA
|
AT-hook transcription factor |
| chr1_-_150765785 | 1.90 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr1_-_206921987 | 1.77 |
ENST00000530505.1
ENST00000442471.4 |
FCMR
|
Fc fragment of IgM receptor |
| chr20_+_33235987 | 1.68 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr12_+_55681711 | 1.67 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr3_-_180679468 | 1.63 |
ENST00000651046.1
ENST00000476379.6 |
CCDC39
|
coiled-coil domain containing 39 |
| chr11_-_72721908 | 1.60 |
ENST00000426523.5
ENST00000429686.5 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_-_121660892 | 1.59 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr4_+_71339014 | 1.50 |
ENST00000340595.4
|
SLC4A4
|
solute carrier family 4 member 4 |
| chr2_+_39665902 | 1.45 |
ENST00000281961.3
ENST00000618232.1 |
TMEM178A
|
transmembrane protein 178A |
| chr2_+_219253243 | 1.44 |
ENST00000490341.3
|
TUBA4B
|
tubulin alpha 4b |
| chr4_+_80335717 | 1.39 |
ENST00000358105.8
ENST00000508675.1 |
CFAP299
|
cilia and flagella associated protein 299 |
| chr8_-_132760548 | 1.34 |
ENST00000519187.5
ENST00000523829.5 ENST00000677595.1 ENST00000356838.7 ENST00000377901.8 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr14_-_22815801 | 1.30 |
ENST00000397532.9
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr1_-_206921867 | 1.29 |
ENST00000628511.2
ENST00000367091.8 |
FCMR
|
Fc fragment of IgM receptor |
| chr14_-_22815856 | 1.28 |
ENST00000554758.1
ENST00000397528.8 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr17_+_4715438 | 1.26 |
ENST00000571206.1
|
ARRB2
|
arrestin beta 2 |
| chr9_+_117704168 | 1.24 |
ENST00000472304.2
ENST00000394487.5 |
TLR4
|
toll like receptor 4 |
| chr2_-_153478753 | 1.22 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr3_-_183555696 | 1.21 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr22_-_38794111 | 1.17 |
ENST00000406622.5
ENST00000216068.9 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein axonemal light chain 4 |
| chr6_+_32844108 | 1.14 |
ENST00000458296.2
ENST00000413039.6 ENST00000412095.1 ENST00000395330.5 |
PSMB8-AS1
PSMB9
|
PSMB8 antisense RNA 1 (head to head) proteasome 20S subunit beta 9 |
| chr19_+_14440254 | 1.14 |
ENST00000342216.8
|
PKN1
|
protein kinase N1 |
| chr11_+_36375978 | 1.13 |
ENST00000378867.7
|
PRR5L
|
proline rich 5 like |
| chr6_+_158649997 | 1.13 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr2_+_120346130 | 1.12 |
ENST00000295228.4
|
INHBB
|
inhibin subunit beta B |
| chr7_+_123601815 | 1.12 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_123601836 | 1.11 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr9_-_4300049 | 1.09 |
ENST00000381971.8
|
GLIS3
|
GLIS family zinc finger 3 |
| chr12_-_94459854 | 1.08 |
ENST00000397809.10
|
CEP83
|
centrosomal protein 83 |
| chr10_+_92831153 | 1.07 |
ENST00000672817.1
|
EXOC6
|
exocyst complex component 6 |
| chr15_-_48963912 | 1.03 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr11_+_72080313 | 1.02 |
ENST00000307198.11
ENST00000538413.6 ENST00000642648.1 ENST00000289488.7 |
ENSG00000284922.2
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing leucine rich transmembrane and O-methyltransferase domain containing |
| chrX_-_133415478 | 1.01 |
ENST00000370828.4
|
GPC4
|
glypican 4 |
| chr14_-_22815421 | 1.00 |
ENST00000674313.1
ENST00000555959.1 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr6_-_32192845 | 1.00 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr3_-_47282752 | 1.00 |
ENST00000456548.5
ENST00000432493.5 ENST00000684063.1 ENST00000444589.6 |
KIF9
|
kinesin family member 9 |
| chr6_-_154356735 | 0.98 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr5_-_160685379 | 0.96 |
ENST00000642502.1
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chrX_+_55452119 | 0.96 |
ENST00000342972.3
|
MAGEH1
|
MAGE family member H1 |
| chr15_+_50182215 | 0.95 |
ENST00000380902.8
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr6_-_32192630 | 0.94 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr22_-_31346317 | 0.93 |
ENST00000266269.10
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr15_+_50182188 | 0.92 |
ENST00000267842.10
|
SLC27A2
|
solute carrier family 27 member 2 |
| chr3_+_48990219 | 0.92 |
ENST00000383729.9
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane |
| chr1_-_183590876 | 0.90 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_-_100894775 | 0.89 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr17_-_36001549 | 0.88 |
ENST00000617897.2
|
CCL15
|
C-C motif chemokine ligand 15 |
| chr16_-_57536543 | 0.86 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr20_+_46118300 | 0.82 |
ENST00000372285.8
ENST00000372276.7 |
CD40
|
CD40 molecule |
| chr2_+_137964446 | 0.82 |
ENST00000280096.5
ENST00000280097.5 |
HNMT
|
histamine N-methyltransferase |
| chr3_+_181711915 | 0.82 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr1_+_108560031 | 0.81 |
ENST00000405454.1
ENST00000370035.8 |
FAM102B
|
family with sequence similarity 102 member B |
| chr8_+_98064522 | 0.81 |
ENST00000545282.1
|
ERICH5
|
glutamate rich 5 |
| chr7_+_114414809 | 0.81 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr8_+_27633863 | 0.80 |
ENST00000337221.8
|
SCARA3
|
scavenger receptor class A member 3 |
| chr1_+_183805105 | 0.80 |
ENST00000360851.4
|
RGL1
|
ral guanine nucleotide dissociation stimulator like 1 |
| chr11_-_72722302 | 0.79 |
ENST00000334211.12
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_-_139084289 | 0.79 |
ENST00000510408.5
ENST00000379549.7 ENST00000358635.7 |
ELF2
|
E74 like ETS transcription factor 2 |
| chr14_+_67533282 | 0.79 |
ENST00000329153.10
|
PLEKHH1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr3_-_47282518 | 0.79 |
ENST00000425853.5
ENST00000452770.6 |
KIF9
|
kinesin family member 9 |
| chr16_+_28494634 | 0.78 |
ENST00000564831.6
ENST00000431282.2 |
APOBR
|
apolipoprotein B receptor |
| chr1_-_157552470 | 0.76 |
ENST00000361835.8
|
FCRL5
|
Fc receptor like 5 |
| chr3_+_113897470 | 0.76 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr5_-_43412323 | 0.76 |
ENST00000361115.4
|
CCL28
|
C-C motif chemokine ligand 28 |
| chr11_-_5441514 | 0.75 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1 |
| chr14_+_44962177 | 0.75 |
ENST00000361462.7
ENST00000361577.7 |
TOGARAM1
|
TOG array regulator of axonemal microtubules 1 |
| chrX_+_129779930 | 0.74 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr19_-_39934626 | 0.74 |
ENST00000616721.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr6_-_6320642 | 0.74 |
ENST00000451619.1
ENST00000264870.8 |
F13A1
|
coagulation factor XIII A chain |
| chr3_-_12158901 | 0.73 |
ENST00000287814.5
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr1_+_39215255 | 0.72 |
ENST00000671089.1
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr7_+_114414997 | 0.72 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr11_+_118304881 | 0.71 |
ENST00000528600.1
|
CD3E
|
CD3e molecule |
| chr5_+_42423433 | 0.71 |
ENST00000230882.9
|
GHR
|
growth hormone receptor |
| chr3_-_11720728 | 0.70 |
ENST00000445411.5
ENST00000273038.7 |
VGLL4
|
vestigial like family member 4 |
| chr11_+_118304721 | 0.70 |
ENST00000361763.9
|
CD3E
|
CD3e molecule |
| chr17_+_7857695 | 0.70 |
ENST00000571846.5
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chrX_-_136780925 | 0.70 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr17_-_48545077 | 0.69 |
ENST00000330070.6
|
HOXB2
|
homeobox B2 |
| chr10_+_112376193 | 0.69 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr1_-_183590439 | 0.69 |
ENST00000367535.8
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr5_-_9546066 | 0.69 |
ENST00000382496.10
ENST00000652226.1 |
SEMA5A
|
semaphorin 5A |
| chr11_-_119340816 | 0.69 |
ENST00000528368.3
|
C1QTNF5
|
C1q and TNF related 5 |
| chr14_+_73537346 | 0.68 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr11_+_6481473 | 0.68 |
ENST00000530751.1
ENST00000254616.11 |
TIMM10B
|
translocase of inner mitochondrial membrane 10B |
| chr12_+_12071396 | 0.67 |
ENST00000396367.5
ENST00000266434.8 |
BCL2L14
|
BCL2 like 14 |
| chr6_-_32843994 | 0.67 |
ENST00000395339.7
ENST00000374882.8 |
PSMB8
|
proteasome 20S subunit beta 8 |
| chr3_-_45842066 | 0.66 |
ENST00000445698.1
ENST00000296135.11 |
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr12_+_12070932 | 0.66 |
ENST00000308721.9
|
BCL2L14
|
BCL2 like 14 |
| chr8_+_98064559 | 0.65 |
ENST00000318528.8
|
ERICH5
|
glutamate rich 5 |
| chr11_-_32435360 | 0.65 |
ENST00000639563.3
|
WT1
|
WT1 transcription factor |
| chr10_+_94089067 | 0.65 |
ENST00000371375.1
ENST00000675218.1 |
PLCE1
|
phospholipase C epsilon 1 |
| chr11_-_6481350 | 0.65 |
ENST00000423813.6
ENST00000614314.4 |
ARFIP2
|
ADP ribosylation factor interacting protein 2 |
| chr20_+_32277626 | 0.64 |
ENST00000375712.4
|
KIF3B
|
kinesin family member 3B |
| chr16_+_4795378 | 0.64 |
ENST00000588606.5
|
SMIM22
|
small integral membrane protein 22 |
| chr11_-_6481304 | 0.64 |
ENST00000254584.6
ENST00000525235.1 ENST00000396777.8 ENST00000445086.6 |
ARFIP2
|
ADP ribosylation factor interacting protein 2 |
| chr11_+_2444986 | 0.64 |
ENST00000155840.12
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr3_-_11720690 | 0.64 |
ENST00000426568.5
|
VGLL4
|
vestigial like family member 4 |
| chr8_-_28490220 | 0.64 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr18_-_55510753 | 0.64 |
ENST00000543082.5
|
TCF4
|
transcription factor 4 |
| chr1_-_183590596 | 0.64 |
ENST00000418089.5
ENST00000413720.5 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr10_+_94089034 | 0.64 |
ENST00000676102.1
ENST00000371385.8 |
PLCE1
|
phospholipase C epsilon 1 |
| chr7_+_102464921 | 0.63 |
ENST00000463739.5
ENST00000292616.10 ENST00000626402.1 |
LRWD1
|
leucine rich repeats and WD repeat domain containing 1 |
| chr6_-_129710145 | 0.63 |
ENST00000368149.3
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr17_-_74712911 | 0.62 |
ENST00000326165.11
ENST00000583937.5 ENST00000301573.13 ENST00000469092.5 |
CD300LF
|
CD300 molecule like family member f |
| chr8_+_80485641 | 0.62 |
ENST00000430430.5
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr16_+_4795357 | 0.62 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chrX_+_17375194 | 0.61 |
ENST00000676302.1
|
NHS
|
NHS actin remodeling regulator |
| chr11_-_47378391 | 0.61 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_150697128 | 0.61 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr12_-_51324652 | 0.61 |
ENST00000544402.5
|
BIN2
|
bridging integrator 2 |
| chr8_+_55879818 | 0.60 |
ENST00000520220.6
ENST00000519728.6 |
LYN
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr12_-_51324164 | 0.60 |
ENST00000615107.6
|
BIN2
|
bridging integrator 2 |
| chr9_+_117704382 | 0.60 |
ENST00000646089.1
ENST00000355622.8 |
ENSG00000285082.2
TLR4
|
novel protein toll like receptor 4 |
| chr12_-_51324138 | 0.60 |
ENST00000452142.7
|
BIN2
|
bridging integrator 2 |
| chr3_+_184337591 | 0.59 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A |
| chr8_+_80486209 | 0.59 |
ENST00000426744.5
ENST00000455036.8 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr1_-_113887375 | 0.59 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2 like 15 |
| chr11_+_6239068 | 0.59 |
ENST00000379936.3
|
CNGA4
|
cyclic nucleotide gated channel subunit alpha 4 |
| chr1_+_171314171 | 0.58 |
ENST00000367749.4
|
FMO4
|
flavin containing dimethylaniline monoxygenase 4 |
| chr12_+_6946468 | 0.58 |
ENST00000543115.5
ENST00000399448.5 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chrX_+_17375230 | 0.58 |
ENST00000380060.7
|
NHS
|
NHS actin remodeling regulator |
| chr1_-_111200633 | 0.58 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr3_-_128123765 | 0.56 |
ENST00000322623.10
|
RUVBL1
|
RuvB like AAA ATPase 1 |
| chr22_-_37149900 | 0.55 |
ENST00000216223.10
|
IL2RB
|
interleukin 2 receptor subunit beta |
| chr12_-_105236074 | 0.55 |
ENST00000551662.5
ENST00000553097.5 ENST00000258530.8 |
APPL2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr9_+_273026 | 0.55 |
ENST00000682249.1
ENST00000453981.5 ENST00000487230.5 ENST00000469391.5 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr19_+_6772699 | 0.55 |
ENST00000602142.6
ENST00000304076.6 ENST00000596764.5 |
VAV1
|
vav guanine nucleotide exchange factor 1 |
| chr21_-_44920855 | 0.55 |
ENST00000397854.7
|
ITGB2
|
integrin subunit beta 2 |
| chr20_+_46118277 | 0.54 |
ENST00000620709.4
|
CD40
|
CD40 molecule |
| chr2_-_101151253 | 0.54 |
ENST00000376840.8
ENST00000409318.2 |
TBC1D8
|
TBC1 domain family member 8 |
| chr9_+_127706975 | 0.54 |
ENST00000373295.7
|
CFAP157
|
cilia and flagella associated protein 157 |
| chr17_-_1645713 | 0.54 |
ENST00000348987.4
ENST00000263071.9 ENST00000571272.5 |
SCARF1
|
scavenger receptor class F member 1 |
| chr11_-_47378527 | 0.54 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr12_-_94459939 | 0.53 |
ENST00000339839.9
ENST00000547575.5 ENST00000546527.1 |
CEP83
|
centrosomal protein 83 |
| chr2_-_148020754 | 0.53 |
ENST00000440042.1
ENST00000536575.5 |
ORC4
|
origin recognition complex subunit 4 |
| chrX_+_96684801 | 0.53 |
ENST00000324765.13
|
DIAPH2
|
diaphanous related formin 2 |
| chr17_-_58328756 | 0.53 |
ENST00000268893.10
ENST00000343736.9 |
TSPOAP1
|
TSPO associated protein 1 |
| chr1_-_157138474 | 0.52 |
ENST00000326786.4
|
ETV3
|
ETS variant transcription factor 3 |
| chr14_-_75980993 | 0.52 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor beta 3 |
| chr12_-_123436664 | 0.52 |
ENST00000280571.10
|
RILPL2
|
Rab interacting lysosomal protein like 2 |
| chr10_-_69409275 | 0.52 |
ENST00000373307.5
|
TACR2
|
tachykinin receptor 2 |
| chr14_-_53954470 | 0.52 |
ENST00000417573.5
|
BMP4
|
bone morphogenetic protein 4 |
| chr10_-_73591330 | 0.51 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chrX_-_55161098 | 0.51 |
ENST00000489298.1
ENST00000477847.6 |
FAM104B
|
family with sequence similarity 104 member B |
| chr12_-_40106026 | 0.51 |
ENST00000280871.9
ENST00000380858.1 |
SLC2A13
|
solute carrier family 2 member 13 |
| chr12_+_25052732 | 0.51 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr2_-_152098810 | 0.50 |
ENST00000636442.1
ENST00000638005.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr10_+_12349533 | 0.50 |
ENST00000619168.5
|
CAMK1D
|
calcium/calmodulin dependent protein kinase ID |
| chr5_+_154445979 | 0.50 |
ENST00000297109.11
|
SAP30L
|
SAP30 like |
| chr19_+_35745613 | 0.50 |
ENST00000222266.2
ENST00000587708.7 |
PSENEN
|
presenilin enhancer, gamma-secretase subunit |
| chr22_-_31346143 | 0.50 |
ENST00000405309.7
ENST00000351933.8 |
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr17_-_31314066 | 0.50 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr3_-_15797930 | 0.50 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr19_+_35745590 | 0.49 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer, gamma-secretase subunit |
| chrX_+_96684712 | 0.49 |
ENST00000373049.8
|
DIAPH2
|
diaphanous related formin 2 |
| chr21_-_39313578 | 0.49 |
ENST00000380800.7
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr12_+_85874287 | 0.49 |
ENST00000551529.5
ENST00000256010.7 |
NTS
|
neurotensin |
| chr2_+_96326204 | 0.49 |
ENST00000420728.1
ENST00000361124.5 |
ITPRIPL1
|
ITPRIP like 1 |
| chr14_+_75985747 | 0.48 |
ENST00000679083.1
ENST00000314067.11 ENST00000238628.10 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 |
| chr3_-_15798184 | 0.47 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr5_-_124744513 | 0.47 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608 |
| chr8_+_27633884 | 0.47 |
ENST00000301904.4
|
SCARA3
|
scavenger receptor class A member 3 |
| chr3_-_114624979 | 0.47 |
ENST00000676079.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_236395394 | 0.47 |
ENST00000359362.6
|
EDARADD
|
EDAR associated death domain |
| chr2_-_148020689 | 0.47 |
ENST00000457954.5
ENST00000392857.10 ENST00000540442.5 ENST00000535373.5 |
ORC4
|
origin recognition complex subunit 4 |
| chr12_+_51391273 | 0.46 |
ENST00000535225.6
ENST00000358657.7 |
SLC4A8
|
solute carrier family 4 member 8 |
| chr3_+_51942323 | 0.46 |
ENST00000431474.6
ENST00000417220.6 ENST00000398755.8 ENST00000471971.6 |
PARP3
|
poly(ADP-ribose) polymerase family member 3 |
| chrX_-_55161156 | 0.46 |
ENST00000472571.2
ENST00000332132.8 ENST00000425133.2 ENST00000358460.8 |
FAM104B
|
family with sequence similarity 104 member B |
| chr16_-_68371005 | 0.46 |
ENST00000574662.1
|
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr11_-_102955705 | 0.46 |
ENST00000615555.4
ENST00000340273.4 ENST00000260302.8 |
MMP13
|
matrix metallopeptidase 13 |
| chr20_-_44210697 | 0.46 |
ENST00000255174.3
|
OSER1
|
oxidative stress responsive serine rich 1 |
| chr19_-_4338786 | 0.46 |
ENST00000601482.1
ENST00000600324.5 ENST00000594605.6 |
STAP2
|
signal transducing adaptor family member 2 |
| chr17_+_40342342 | 0.46 |
ENST00000394081.7
|
RARA
|
retinoic acid receptor alpha |
| chr11_+_103109522 | 0.46 |
ENST00000334267.11
|
DYNC2H1
|
dynein cytoplasmic 2 heavy chain 1 |
| chrX_-_10833643 | 0.46 |
ENST00000380785.5
ENST00000380787.5 |
MID1
|
midline 1 |
| chr8_+_73991345 | 0.45 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_+_49020443 | 0.45 |
ENST00000326912.8
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr1_-_100894818 | 0.45 |
ENST00000370114.8
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr2_+_96537254 | 0.45 |
ENST00000454558.2
|
ARID5A
|
AT-rich interaction domain 5A |
| chr3_-_114624921 | 0.45 |
ENST00000393785.6
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_+_76948500 | 0.45 |
ENST00000377462.6
|
ACOD1
|
aconitate decarboxylase 1 |
| chr3_+_171843337 | 0.45 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr3_+_37020333 | 0.45 |
ENST00000616768.5
|
MLH1
|
mutL homolog 1 |
| chr17_-_37542361 | 0.44 |
ENST00000614196.1
|
SYNRG
|
synergin gamma |
| chr12_+_25052634 | 0.44 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_-_100895132 | 0.44 |
ENST00000535414.5
|
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr7_+_48171451 | 0.44 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr2_-_173964069 | 0.43 |
ENST00000652005.2
|
SP3
|
Sp3 transcription factor |
| chr3_-_114624193 | 0.43 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_149333407 | 0.43 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chrX_+_96684638 | 0.43 |
ENST00000355827.8
ENST00000373061.7 |
DIAPH2
|
diaphanous related formin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 1.8 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.6 | 2.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.5 | 3.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.5 | 1.5 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.5 | 1.9 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.4 | 1.7 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.4 | 1.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.4 | 1.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 1.0 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.3 | 1.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 1.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 0.9 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.3 | 1.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.3 | 1.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 1.0 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.2 | 0.7 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 0.2 | 1.4 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.2 | 0.7 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 1.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.6 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) mast cell proliferation(GO:0070662) |
| 0.2 | 0.8 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 0.6 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.2 | 0.8 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.2 | 1.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.7 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 1.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.6 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.4 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.3 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.5 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.8 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 1.6 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 1.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.5 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 1.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.1 | 0.2 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.8 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.5 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.4 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.3 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.8 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.5 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 0.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.2 | GO:2000395 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 1.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.2 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) L-arginine transmembrane transport(GO:1903400) |
| 0.1 | 0.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.5 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 1.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.0 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.6 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.6 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 2.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.3 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.3 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 1.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.4 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.8 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 1.0 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 2.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.8 | GO:2000352 | negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:2000503 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.4 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 2.3 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.6 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.3 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.5 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.8 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:0032780 | protein import into mitochondrial matrix(GO:0030150) negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.3 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.7 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.0 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 2.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 2.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 0.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.2 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 2.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.0 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.4 | 2.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 2.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 1.0 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.2 | 3.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.2 | 2.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.5 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.4 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.8 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 2.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.0 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.9 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 1.0 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 4.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.7 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 2.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.7 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 3.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 1.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 2.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 4.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |