Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
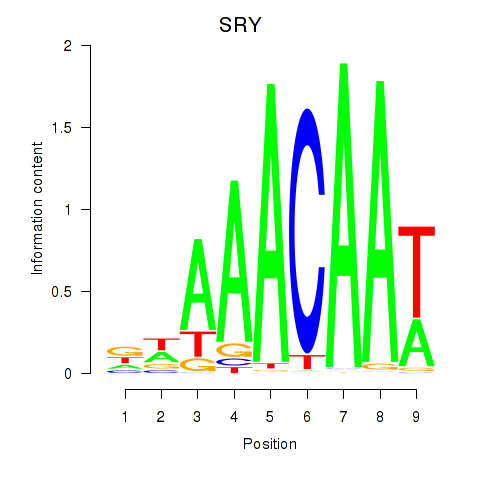
Results for SRY
Z-value: 0.76
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.8 | SRY |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SRY | hg38_v1_chrY_-_2787676_2787687 | -0.32 | 8.6e-02 | Click! |
Activity profile of SRY motif
Sorted Z-values of SRY motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_38990824 | 4.00 |
ENST00000379631.9
|
STOML3
|
stomatin like 3 |
| chr13_-_38990856 | 3.93 |
ENST00000423210.1
|
STOML3
|
stomatin like 3 |
| chr20_-_3781440 | 2.85 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr6_-_32589833 | 2.83 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr4_+_164754045 | 2.63 |
ENST00000515485.5
|
SMIM31
|
small integral membrane protein 31 |
| chr6_+_162727941 | 2.33 |
ENST00000366888.6
|
PACRG
|
parkin coregulated |
| chr17_+_38705482 | 2.20 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chr17_+_17972813 | 1.95 |
ENST00000582416.5
ENST00000313838.12 ENST00000399187.6 ENST00000581264.5 ENST00000479684.2 ENST00000584166.5 ENST00000585108.5 ENST00000399182.5 ENST00000579977.1 |
DRC3
|
dynein regulatory complex subunit 3 |
| chr7_+_114414809 | 1.91 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr4_+_164754116 | 1.88 |
ENST00000507311.1
|
SMIM31
|
small integral membrane protein 31 |
| chr1_-_217137748 | 1.54 |
ENST00000493603.5
ENST00000366940.6 |
ESRRG
|
estrogen related receptor gamma |
| chr8_-_71361860 | 1.44 |
ENST00000303824.11
ENST00000645451.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_-_86337654 | 1.43 |
ENST00000165698.9
|
REEP1
|
receptor accessory protein 1 |
| chr2_-_86337617 | 1.43 |
ENST00000538924.7
ENST00000535845.6 |
REEP1
|
receptor accessory protein 1 |
| chr14_-_91947654 | 1.37 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr14_-_91947383 | 1.31 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr1_-_39901996 | 1.30 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr1_-_217076889 | 1.22 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr7_-_117873420 | 1.19 |
ENST00000160373.8
|
CTTNBP2
|
cortactin binding protein 2 |
| chr1_+_61081728 | 1.17 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr17_-_58328756 | 1.15 |
ENST00000268893.10
ENST00000343736.9 |
TSPOAP1
|
TSPO associated protein 1 |
| chr10_-_60572599 | 1.12 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr6_-_152168349 | 1.10 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr6_-_152168291 | 1.07 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr20_-_21514046 | 1.03 |
ENST00000377142.5
|
NKX2-2
|
NK2 homeobox 2 |
| chr8_-_71547626 | 1.00 |
ENST00000647540.1
ENST00000644229.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr14_-_91946989 | 0.95 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr14_-_22819721 | 0.94 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr2_-_74421560 | 0.90 |
ENST00000612891.4
ENST00000640331.1 ENST00000684111.1 |
C2orf81
|
chromosome 2 open reading frame 81 |
| chr9_+_2110354 | 0.88 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_+_13023958 | 0.88 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr2_-_156342348 | 0.85 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr10_+_113125536 | 0.84 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr10_-_60141004 | 0.83 |
ENST00000355288.6
|
ANK3
|
ankyrin 3 |
| chr1_-_39901861 | 0.83 |
ENST00000372816.3
ENST00000372815.1 |
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr11_+_36296281 | 0.82 |
ENST00000530639.6
|
PRR5L
|
proline rich 5 like |
| chr6_-_31684040 | 0.82 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr3_-_11568764 | 0.81 |
ENST00000424529.6
|
VGLL4
|
vestigial like family member 4 |
| chr14_+_91114667 | 0.81 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr7_-_78771265 | 0.80 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_140868945 | 0.79 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr22_-_31345770 | 0.77 |
ENST00000215919.3
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr18_-_55321986 | 0.77 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr14_-_89417148 | 0.76 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chr1_+_33256479 | 0.75 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr2_+_190408324 | 0.75 |
ENST00000417958.5
ENST00000432036.5 ENST00000392328.6 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr9_-_122227525 | 0.74 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr14_+_91114431 | 0.74 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chr11_-_85686123 | 0.73 |
ENST00000316398.5
|
CCDC89
|
coiled-coil domain containing 89 |
| chr14_+_91114364 | 0.72 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr10_-_60389833 | 0.72 |
ENST00000280772.7
|
ANK3
|
ankyrin 3 |
| chr17_+_9021501 | 0.72 |
ENST00000173229.7
|
NTN1
|
netrin 1 |
| chr5_-_111756245 | 0.72 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr1_+_61082553 | 0.70 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr10_+_97319250 | 0.70 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1 |
| chr4_+_94974984 | 0.69 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr14_-_52552493 | 0.68 |
ENST00000281741.9
ENST00000557374.1 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr9_-_14314132 | 0.68 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chr9_-_134068518 | 0.67 |
ENST00000371834.6
|
BRD3
|
bromodomain containing 3 |
| chr6_+_122399536 | 0.67 |
ENST00000452194.5
|
HSF2
|
heat shock transcription factor 2 |
| chr12_-_85836372 | 0.67 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr18_-_55589770 | 0.65 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr6_+_122399621 | 0.65 |
ENST00000368455.9
|
HSF2
|
heat shock transcription factor 2 |
| chr16_-_68448491 | 0.65 |
ENST00000561749.1
ENST00000219334.10 |
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr6_-_87095059 | 0.65 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr9_-_71768386 | 0.63 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr10_-_97334698 | 0.60 |
ENST00000371019.4
|
FRAT2
|
FRAT regulator of WNT signaling pathway 2 |
| chr18_+_75210789 | 0.60 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr11_-_16356538 | 0.58 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr18_-_55351977 | 0.58 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr1_+_87328860 | 0.58 |
ENST00000370544.10
|
LMO4
|
LIM domain only 4 |
| chr1_-_56579555 | 0.57 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr2_+_86441341 | 0.57 |
ENST00000312912.10
ENST00000409064.5 |
KDM3A
|
lysine demethylase 3A |
| chr18_-_55321640 | 0.57 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr18_+_75210755 | 0.56 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr2_-_71227055 | 0.56 |
ENST00000244221.9
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr7_+_114414997 | 0.56 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr4_-_104494882 | 0.55 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4 |
| chr18_-_55322215 | 0.53 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr9_+_27109393 | 0.53 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr5_-_147081428 | 0.52 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr4_+_41612892 | 0.52 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_13795434 | 0.52 |
ENST00000254323.6
|
ZSWIM4
|
zinc finger SWIM-type containing 4 |
| chr17_+_59331633 | 0.50 |
ENST00000312655.9
|
YPEL2
|
yippee like 2 |
| chr18_-_55589795 | 0.50 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr6_+_156777366 | 0.50 |
ENST00000636930.2
|
ARID1B
|
AT-rich interaction domain 1B |
| chr1_+_40374648 | 0.50 |
ENST00000372708.5
|
SMAP2
|
small ArfGAP2 |
| chr9_-_127980976 | 0.50 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr12_-_122500924 | 0.48 |
ENST00000633063.3
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr21_-_17819343 | 0.48 |
ENST00000284881.9
|
C21orf91
|
chromosome 21 open reading frame 91 |
| chr18_-_55589836 | 0.48 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr18_-_55635948 | 0.47 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr4_-_173530219 | 0.47 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr10_-_50279715 | 0.47 |
ENST00000395526.9
|
ASAH2
|
N-acylsphingosine amidohydrolase 2 |
| chr12_-_122500947 | 0.46 |
ENST00000672018.1
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr5_-_147056010 | 0.46 |
ENST00000394414.5
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr3_-_112641292 | 0.45 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr6_+_156777882 | 0.44 |
ENST00000350026.10
ENST00000647938.1 ENST00000674298.1 |
ARID1B
|
AT-rich interaction domain 1B |
| chr5_-_147055968 | 0.44 |
ENST00000336640.10
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr14_+_91114388 | 0.43 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chr12_-_122500520 | 0.41 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr7_+_30145789 | 0.41 |
ENST00000324489.5
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog |
| chr11_-_95910824 | 0.41 |
ENST00000674528.1
ENST00000675477.1 ENST00000675636.1 |
MTMR2
|
myotubularin related protein 2 |
| chr6_+_138773747 | 0.40 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr3_-_142149515 | 0.40 |
ENST00000475734.5
ENST00000467072.5 ENST00000489671.6 |
TFDP2
|
transcription factor Dp-2 |
| chr19_+_13024917 | 0.39 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X |
| chr12_-_24562438 | 0.39 |
ENST00000646273.1
ENST00000659413.1 ENST00000446891.7 |
SOX5
|
SRY-box transcription factor 5 |
| chr7_-_78771108 | 0.39 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_-_24672654 | 0.39 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr18_-_55302613 | 0.38 |
ENST00000561831.7
|
TCF4
|
transcription factor 4 |
| chr14_+_91114026 | 0.38 |
ENST00000521081.5
ENST00000520328.5 ENST00000524232.5 ENST00000522170.5 ENST00000256324.15 ENST00000519950.5 ENST00000523879.5 ENST00000521077.6 ENST00000518665.6 |
DGLUCY
|
D-glutamate cyclase |
| chr10_+_91162958 | 0.38 |
ENST00000614189.4
|
PCGF5
|
polycomb group ring finger 5 |
| chr11_-_95910748 | 0.38 |
ENST00000675933.1
|
MTMR2
|
myotubularin related protein 2 |
| chr11_-_95910665 | 0.38 |
ENST00000674610.1
ENST00000675660.1 |
MTMR2
|
myotubularin related protein 2 |
| chr20_+_10435283 | 0.37 |
ENST00000334534.10
|
SLX4IP
|
SLX4 interacting protein |
| chr1_+_40709475 | 0.37 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr11_-_44950151 | 0.37 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr8_+_109334317 | 0.36 |
ENST00000521662.5
ENST00000520147.5 ENST00000521688.6 |
ENY2
|
ENY2 transcription and export complex 2 subunit |
| chr2_-_159616442 | 0.36 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr21_-_17819386 | 0.35 |
ENST00000400559.7
ENST00000400558.7 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr11_-_115504389 | 0.35 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr1_-_52552994 | 0.34 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr16_+_2537997 | 0.34 |
ENST00000441549.7
ENST00000268673.11 ENST00000342085.9 ENST00000389224.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase 1 |
| chr5_-_147081462 | 0.34 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr18_+_58341038 | 0.34 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_107688492 | 0.33 |
ENST00000415432.6
|
VAV3
|
vav guanine nucleotide exchange factor 3 |
| chr9_-_134068012 | 0.32 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr9_-_14314567 | 0.32 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr10_+_110207587 | 0.32 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr7_-_93226449 | 0.31 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr2_+_27442365 | 0.31 |
ENST00000543753.5
ENST00000288873.7 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr4_+_169660062 | 0.30 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr10_-_48274567 | 0.30 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr9_+_2159672 | 0.30 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_177164673 | 0.30 |
ENST00000424564.2
ENST00000361833.7 |
ASTN1
|
astrotactin 1 |
| chr15_-_53733103 | 0.29 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr2_-_231125032 | 0.28 |
ENST00000258400.4
|
HTR2B
|
5-hydroxytryptamine receptor 2B |
| chr8_-_56211257 | 0.28 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chrX_+_15507302 | 0.28 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr2_+_27442421 | 0.28 |
ENST00000407293.5
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr11_+_31816266 | 0.27 |
ENST00000644607.1
ENST00000646221.1 ENST00000643671.1 ENST00000643931.1 ENST00000642614.1 ENST00000642818.1 ENST00000645848.1 ENST00000506388.2 ENST00000645824.1 ENST00000532942.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr8_-_143986425 | 0.27 |
ENST00000313059.9
ENST00000524918.5 ENST00000313028.12 ENST00000525773.5 |
PARP10
|
poly(ADP-ribose) polymerase family member 10 |
| chr6_-_116060859 | 0.26 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr1_+_164559739 | 0.25 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr5_+_141177790 | 0.25 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr16_-_3880678 | 0.25 |
ENST00000262367.10
|
CREBBP
|
CREB binding protein |
| chr9_-_14313843 | 0.24 |
ENST00000636063.1
ENST00000380921.3 ENST00000622520.1 ENST00000380959.7 |
NFIB
|
nuclear factor I B |
| chr10_-_32378720 | 0.24 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr9_-_14314519 | 0.24 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr13_-_40771105 | 0.23 |
ENST00000323563.8
|
MRPS31
|
mitochondrial ribosomal protein S31 |
| chr3_+_142623386 | 0.23 |
ENST00000337777.7
ENST00000497199.5 |
PLS1
|
plastin 1 |
| chr2_+_108588286 | 0.23 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr1_+_43389889 | 0.23 |
ENST00000562955.2
ENST00000634258.3 |
SZT2
|
SZT2 subunit of KICSTOR complex |
| chr12_-_10098977 | 0.23 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr9_-_37465402 | 0.23 |
ENST00000307750.5
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr19_-_47231191 | 0.22 |
ENST00000439096.3
|
BBC3
|
BCL2 binding component 3 |
| chr1_+_40709316 | 0.22 |
ENST00000372652.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr12_+_389334 | 0.22 |
ENST00000540180.5
ENST00000422000.5 ENST00000535052.5 |
CCDC77
|
coiled-coil domain containing 77 |
| chr3_-_58666765 | 0.21 |
ENST00000358781.7
|
FAM3D
|
FAM3 metabolism regulating signaling molecule D |
| chr1_+_147902789 | 0.21 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr3_+_41194741 | 0.21 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr12_-_389249 | 0.21 |
ENST00000535014.5
ENST00000543507.1 ENST00000544760.1 ENST00000399788.7 |
KDM5A
|
lysine demethylase 5A |
| chr16_+_28846674 | 0.21 |
ENST00000322610.12
|
SH2B1
|
SH2B adaptor protein 1 |
| chr16_-_46763237 | 0.21 |
ENST00000536476.5
|
MYLK3
|
myosin light chain kinase 3 |
| chr7_-_19773569 | 0.21 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr14_+_52553273 | 0.20 |
ENST00000542169.6
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr14_+_52552830 | 0.20 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr1_+_164559766 | 0.20 |
ENST00000367897.5
ENST00000559240.5 |
PBX1
|
PBX homeobox 1 |
| chr20_-_56497608 | 0.20 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr17_-_75515509 | 0.20 |
ENST00000321617.8
|
CASKIN2
|
CASK interacting protein 2 |
| chr8_-_42501224 | 0.20 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr9_-_14313642 | 0.20 |
ENST00000637742.1
|
NFIB
|
nuclear factor I B |
| chr1_+_235327344 | 0.19 |
ENST00000488594.5
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_+_84164684 | 0.19 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr13_-_40666600 | 0.19 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr3_-_62373015 | 0.19 |
ENST00000475839.1
|
FEZF2
|
FEZ family zinc finger 2 |
| chr8_-_86743626 | 0.19 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr15_+_76336755 | 0.19 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr20_+_33562365 | 0.19 |
ENST00000346541.7
ENST00000397800.5 ENST00000492345.5 |
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr2_+_86720282 | 0.18 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr9_-_95509241 | 0.18 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chr5_+_111071710 | 0.18 |
ENST00000344895.4
|
TSLP
|
thymic stromal lymphopoietin |
| chr11_-_30586866 | 0.18 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr5_+_163505564 | 0.18 |
ENST00000421814.6
ENST00000518095.5 ENST00000321757.11 |
MAT2B
|
methionine adenosyltransferase 2B |
| chr12_+_11649666 | 0.17 |
ENST00000396373.9
|
ETV6
|
ETS variant transcription factor 6 |
| chr5_+_139341875 | 0.17 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr22_+_19723525 | 0.17 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr2_+_134838610 | 0.16 |
ENST00000356140.10
ENST00000392928.5 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_+_94995919 | 0.16 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr11_+_112961480 | 0.16 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr16_+_30395400 | 0.16 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr6_-_37697875 | 0.16 |
ENST00000434837.8
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr12_-_86256267 | 0.15 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr7_-_149028452 | 0.15 |
ENST00000413966.1
ENST00000652332.1 |
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr5_+_139342442 | 0.15 |
ENST00000394795.6
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_+_58427425 | 0.15 |
ENST00000619722.5
ENST00000423743.7 |
KIAA0586
|
KIAA0586 |
| chr7_+_29479712 | 0.15 |
ENST00000412711.6
|
CHN2
|
chimerin 2 |
| chr2_+_108588453 | 0.15 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr22_-_49827512 | 0.15 |
ENST00000404760.5
|
BRD1
|
bromodomain containing 1 |
| chr13_-_99258366 | 0.15 |
ENST00000397470.5
ENST00000397473.7 |
GPR18
|
G protein-coupled receptor 18 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0021758 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.7 | 2.8 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.7 | 2.7 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 3.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 0.6 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 0.8 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 2.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 2.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 2.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.6 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.2 | 1.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.9 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.9 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 2.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.2 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 1.0 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 1.2 | GO:0031642 | negative regulation of myelination(GO:0031642) positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 1.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.1 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 1.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0008354 | germ cell migration(GO:0008354) Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 1.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 2.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 3.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 4.6 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0032599 | B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 2.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 7.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 1.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.3 | 2.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 4.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 2.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.0 | 3.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 3.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 4.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 3.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |