Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
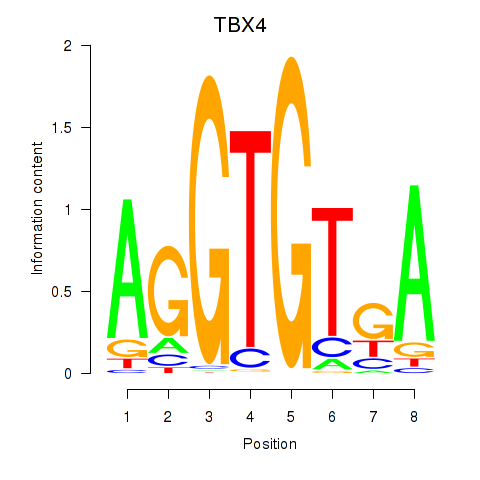
Results for TBX4
Z-value: 0.39
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.11 | TBX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX4 | hg38_v1_chr17_+_61452378_61452437 | -0.09 | 6.5e-01 | Click! |
Activity profile of TBX4 motif
Sorted Z-values of TBX4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_76626127 | 0.42 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr14_+_75280078 | 0.34 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr11_+_61680373 | 0.31 |
ENST00000257215.10
|
DAGLA
|
diacylglycerol lipase alpha |
| chr1_+_145927105 | 0.28 |
ENST00000437797.5
ENST00000601726.3 ENST00000599626.5 ENST00000599147.5 ENST00000595494.5 ENST00000595518.5 ENST00000597144.5 ENST00000599469.5 ENST00000598354.5 ENST00000598103.5 ENST00000600340.5 ENST00000630257.2 ENST00000625258.1 |
LIX1L-AS1
ENSG00000280778.1
|
LIX1L antisense RNA 1 novel protein, lncRNA-POLR3GL readthrough |
| chr14_+_75278820 | 0.28 |
ENST00000554617.1
ENST00000554212.5 ENST00000535987.5 ENST00000303562.9 ENST00000555242.1 |
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr11_-_34513750 | 0.27 |
ENST00000532417.1
|
ELF5
|
E74 like ETS transcription factor 5 |
| chrX_+_16786421 | 0.25 |
ENST00000398155.4
ENST00000380122.10 |
TXLNG
|
taxilin gamma |
| chr14_+_66824439 | 0.25 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr17_-_42181081 | 0.24 |
ENST00000607371.5
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr17_-_42181116 | 0.24 |
ENST00000264661.4
|
KCNH4
|
potassium voltage-gated channel subfamily H member 4 |
| chr8_-_94895195 | 0.22 |
ENST00000308108.9
ENST00000396133.7 |
CCNE2
|
cyclin E2 |
| chr4_+_8580387 | 0.22 |
ENST00000382487.5
|
GPR78
|
G protein-coupled receptor 78 |
| chr14_+_78170336 | 0.22 |
ENST00000634499.2
ENST00000335750.7 |
NRXN3
|
neurexin 3 |
| chr3_+_63819368 | 0.19 |
ENST00000616659.1
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr12_-_52703522 | 0.18 |
ENST00000341809.8
|
KRT77
|
keratin 77 |
| chr3_+_63819341 | 0.18 |
ENST00000647022.1
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr12_-_7695752 | 0.17 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3 |
| chr17_-_41060109 | 0.17 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr13_-_29595670 | 0.16 |
ENST00000380752.10
|
SLC7A1
|
solute carrier family 7 member 1 |
| chr17_-_41065879 | 0.16 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr19_+_38789198 | 0.16 |
ENST00000314980.5
|
LGALS7B
|
galectin 7B |
| chr5_+_150660841 | 0.15 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3 |
| chr15_+_92393841 | 0.15 |
ENST00000268164.8
ENST00000539113.5 ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr19_+_35031263 | 0.14 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr16_+_69105636 | 0.14 |
ENST00000569188.6
|
HAS3
|
hyaluronan synthase 3 |
| chr5_+_114362286 | 0.14 |
ENST00000610748.4
ENST00000264773.7 |
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr5_+_114362043 | 0.14 |
ENST00000673685.1
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr14_+_78403686 | 0.13 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr1_+_26410809 | 0.13 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr12_+_18738102 | 0.13 |
ENST00000317658.5
|
CAPZA3
|
capping actin protein of muscle Z-line subunit alpha 3 |
| chr2_+_176107272 | 0.13 |
ENST00000249504.7
|
HOXD11
|
homeobox D11 |
| chr5_+_150661243 | 0.13 |
ENST00000517768.6
|
MYOZ3
|
myozenin 3 |
| chr8_-_98942557 | 0.12 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3 |
| chr2_-_40453438 | 0.12 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr17_-_45425620 | 0.12 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr12_+_65278919 | 0.12 |
ENST00000538045.5
ENST00000642411.1 ENST00000535239.5 ENST00000614640.4 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr14_+_93430927 | 0.12 |
ENST00000393151.6
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr21_+_29130630 | 0.12 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_-_86068929 | 0.11 |
ENST00000630913.2
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_175005357 | 0.11 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr4_+_139301478 | 0.11 |
ENST00000296543.10
ENST00000398947.1 |
NAA15
|
N-alpha-acetyltransferase 15, NatA auxiliary subunit |
| chr22_+_40045451 | 0.11 |
ENST00000402203.5
|
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr2_+_209771972 | 0.11 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr3_+_167735704 | 0.11 |
ENST00000446050.7
ENST00000295777.9 ENST00000472747.2 |
SERPINI1
|
serpin family I member 1 |
| chr7_-_27102669 | 0.11 |
ENST00000222718.7
|
HOXA2
|
homeobox A2 |
| chr2_-_199457931 | 0.11 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr1_-_11848345 | 0.11 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr1_-_11847772 | 0.11 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr2_-_197675578 | 0.11 |
ENST00000295049.9
|
RFTN2
|
raftlin family member 2 |
| chr11_-_34513785 | 0.10 |
ENST00000257832.7
ENST00000429939.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr14_+_93430853 | 0.10 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr8_+_32548210 | 0.10 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr5_+_75511756 | 0.10 |
ENST00000241436.8
|
POLK
|
DNA polymerase kappa |
| chr15_+_60004305 | 0.10 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr8_+_32548267 | 0.10 |
ENST00000356819.7
|
NRG1
|
neuregulin 1 |
| chr11_-_86068743 | 0.10 |
ENST00000356360.9
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_+_32548303 | 0.10 |
ENST00000650967.1
|
NRG1
|
neuregulin 1 |
| chr7_+_143316105 | 0.10 |
ENST00000343257.7
ENST00000650516.1 |
CLCN1
|
chloride voltage-gated channel 1 |
| chr12_-_116276759 | 0.10 |
ENST00000548743.2
|
MED13L
|
mediator complex subunit 13L |
| chr8_+_32647080 | 0.09 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr10_+_93073873 | 0.09 |
ENST00000224356.5
|
CYP26A1
|
cytochrome P450 family 26 subfamily A member 1 |
| chr1_+_15736251 | 0.09 |
ENST00000294454.6
|
SLC25A34
|
solute carrier family 25 member 34 |
| chr15_+_75724034 | 0.09 |
ENST00000332145.3
|
ODF3L1
|
outer dense fiber of sperm tails 3 like 1 |
| chr9_+_34990250 | 0.09 |
ENST00000454002.6
ENST00000545841.5 |
DNAJB5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr20_+_46029165 | 0.09 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr6_+_39792298 | 0.09 |
ENST00000633794.1
ENST00000274867.9 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr2_+_209771814 | 0.09 |
ENST00000673951.1
ENST00000673920.1 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr15_-_58065870 | 0.08 |
ENST00000537372.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr14_+_52730154 | 0.08 |
ENST00000354586.5
ENST00000442123.6 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr1_-_182672232 | 0.08 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr20_+_46029206 | 0.08 |
ENST00000243964.7
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr15_-_58065734 | 0.08 |
ENST00000347587.7
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr5_-_79514127 | 0.08 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr3_+_134795277 | 0.08 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr15_-_58065703 | 0.08 |
ENST00000249750.9
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr1_-_110606009 | 0.08 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr11_+_18322253 | 0.08 |
ENST00000453096.6
|
GTF2H1
|
general transcription factor IIH subunit 1 |
| chr19_+_8444967 | 0.08 |
ENST00000600092.5
ENST00000325495.9 ENST00000620401.4 ENST00000594907.5 ENST00000596984.5 ENST00000601645.5 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr15_+_89776326 | 0.08 |
ENST00000341735.5
|
MESP2
|
mesoderm posterior bHLH transcription factor 2 |
| chr5_+_139795795 | 0.08 |
ENST00000274710.4
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr2_+_37231633 | 0.08 |
ENST00000002125.9
ENST00000336237.10 ENST00000431821.5 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chrX_+_124346525 | 0.08 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr11_-_7796942 | 0.08 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr1_+_206507546 | 0.08 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr1_+_158355894 | 0.08 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chrX_+_70423031 | 0.07 |
ENST00000453994.6
ENST00000538649.5 ENST00000536730.5 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr15_-_82647503 | 0.07 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chrX_+_70423301 | 0.07 |
ENST00000374382.4
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr2_-_61471062 | 0.07 |
ENST00000398571.7
|
USP34
|
ubiquitin specific peptidase 34 |
| chr22_-_40819324 | 0.07 |
ENST00000435456.7
ENST00000434185.1 ENST00000544408.5 |
SLC25A17
|
solute carrier family 25 member 17 |
| chr1_+_165827574 | 0.07 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr8_+_32646838 | 0.07 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr1_+_165827786 | 0.07 |
ENST00000642653.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chrX_+_124346544 | 0.07 |
ENST00000371139.9
|
SH2D1A
|
SH2 domain containing 1A |
| chr2_+_202073282 | 0.07 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr4_-_101347492 | 0.07 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr2_+_202073249 | 0.06 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr2_+_37231798 | 0.06 |
ENST00000439218.5
ENST00000432075.1 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr10_-_75109085 | 0.06 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr4_-_101347471 | 0.06 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr11_-_16356538 | 0.06 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr1_+_95117324 | 0.06 |
ENST00000370203.9
ENST00000456991.5 |
TLCD4
|
TLC domain containing 4 |
| chr1_+_206507520 | 0.06 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chrX_+_71223216 | 0.06 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1 |
| chr12_+_57460127 | 0.06 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr12_-_18738006 | 0.05 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chrX_+_110002635 | 0.05 |
ENST00000372072.7
|
TMEM164
|
transmembrane protein 164 |
| chr12_+_65279092 | 0.05 |
ENST00000646299.1
|
MSRB3
|
methionine sulfoxide reductase B3 |
| chr12_-_89630552 | 0.05 |
ENST00000393164.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr8_+_55102448 | 0.05 |
ENST00000622811.1
|
XKR4
|
XK related 4 |
| chr12_+_65278643 | 0.05 |
ENST00000355192.8
ENST00000308259.10 ENST00000540804.5 ENST00000535664.5 ENST00000541189.5 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr3_+_52777580 | 0.05 |
ENST00000273283.7
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr11_+_18322541 | 0.05 |
ENST00000534641.5
ENST00000265963.9 ENST00000525831.5 |
GTF2H1
|
general transcription factor IIH subunit 1 |
| chr5_+_60945193 | 0.05 |
ENST00000296597.10
ENST00000511107.1 ENST00000677932.1 ENST00000502658.1 |
NDUFAF2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr1_+_61203496 | 0.05 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A |
| chr4_-_101347327 | 0.05 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr19_-_1863497 | 0.05 |
ENST00000617223.1
ENST00000250916.6 |
KLF16
|
Kruppel like factor 16 |
| chr15_+_36702009 | 0.05 |
ENST00000562489.1
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr1_-_182671853 | 0.05 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr1_-_182671902 | 0.05 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8 |
| chrX_+_124346571 | 0.04 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr2_-_110534010 | 0.04 |
ENST00000437167.1
|
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr3_-_49422429 | 0.04 |
ENST00000637682.1
ENST00000427987.6 ENST00000636199.1 ENST00000638063.1 ENST00000636865.1 ENST00000458307.6 ENST00000273588.9 ENST00000635808.1 ENST00000636522.1 ENST00000636597.1 ENST00000538581.6 ENST00000395338.7 ENST00000462048.2 |
AMT
|
aminomethyltransferase |
| chr12_-_102478539 | 0.04 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr21_-_25734887 | 0.04 |
ENST00000400094.5
ENST00000284971.8 |
ATP5PF
|
ATP synthase peripheral stalk subunit F6 |
| chr12_+_103930600 | 0.04 |
ENST00000680316.1
ENST00000679861.1 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
| chr7_-_120857124 | 0.04 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr12_+_65279445 | 0.04 |
ENST00000642404.1
|
MSRB3
|
methionine sulfoxide reductase B3 |
| chr11_+_64306227 | 0.04 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chrX_-_134796256 | 0.04 |
ENST00000486347.5
|
PABIR2
|
PABIR family member 2 |
| chr17_-_63911236 | 0.03 |
ENST00000438387.6
ENST00000346606.10 ENST00000309894.6 ENST00000561003.5 ENST00000450719.3 ENST00000259003.14 |
CSHL1
|
chorionic somatomammotropin hormone like 1 |
| chr11_+_6481473 | 0.03 |
ENST00000530751.1
ENST00000254616.11 |
TIMM10B
|
translocase of inner mitochondrial membrane 10B |
| chr1_+_154405326 | 0.03 |
ENST00000368485.8
|
IL6R
|
interleukin 6 receptor |
| chr9_+_133534697 | 0.03 |
ENST00000651351.2
|
ADAMTSL2
|
ADAMTS like 2 |
| chr21_+_25734948 | 0.03 |
ENST00000400075.4
|
GABPA
|
GA binding protein transcription factor subunit alpha |
| chr2_+_218607914 | 0.03 |
ENST00000417849.5
|
PLCD4
|
phospholipase C delta 4 |
| chr9_+_133534807 | 0.03 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS like 2 |
| chr11_-_128867364 | 0.03 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr8_+_737595 | 0.03 |
ENST00000637795.2
|
DLGAP2
|
DLG associated protein 2 |
| chr2_-_37231549 | 0.03 |
ENST00000234170.10
|
CEBPZ
|
CCAAT enhancer binding protein zeta |
| chr11_-_65862026 | 0.03 |
ENST00000532134.5
|
CFL1
|
cofilin 1 |
| chr11_-_128867268 | 0.03 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr4_-_139301204 | 0.03 |
ENST00000505036.5
ENST00000539002.5 ENST00000544855.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr21_-_25735026 | 0.03 |
ENST00000400099.5
ENST00000457143.6 |
ATP5PF
|
ATP synthase peripheral stalk subunit F6 |
| chr12_+_103930332 | 0.03 |
ENST00000681861.1
ENST00000550595.2 ENST00000680762.1 ENST00000614327.2 ENST00000681949.1 ENST00000299767.10 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
| chr7_+_107580215 | 0.03 |
ENST00000465919.5
ENST00000005259.9 ENST00000445771.6 ENST00000479917.5 ENST00000421217.5 ENST00000457837.5 |
BCAP29
|
B cell receptor associated protein 29 |
| chr10_-_97771954 | 0.03 |
ENST00000266066.4
|
SFRP5
|
secreted frizzled related protein 5 |
| chr1_+_154405193 | 0.02 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr7_+_107580454 | 0.02 |
ENST00000379117.6
ENST00000473124.1 |
BCAP29
|
B cell receptor associated protein 29 |
| chr2_+_170715317 | 0.02 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor |
| chrX_-_19670983 | 0.02 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr11_-_111879425 | 0.02 |
ENST00000622211.4
|
ENSG00000258529.5
|
novel protein |
| chr2_-_2326378 | 0.02 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr7_+_27242796 | 0.02 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr2_+_152335163 | 0.02 |
ENST00000288670.14
|
FMNL2
|
formin like 2 |
| chr20_-_22584547 | 0.02 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr1_+_26787926 | 0.02 |
ENST00000674202.1
ENST00000674222.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr1_+_26788166 | 0.01 |
ENST00000374145.6
ENST00000431541.6 ENST00000674273.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr6_-_13487593 | 0.01 |
ENST00000379287.4
ENST00000603223.1 |
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr16_-_18926408 | 0.01 |
ENST00000446231.7
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr11_-_18322122 | 0.01 |
ENST00000349215.8
ENST00000396253.7 ENST00000438420.6 |
HPS5
|
HPS5 biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr2_+_102473219 | 0.01 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr13_+_114281577 | 0.01 |
ENST00000375299.8
ENST00000492270.1 ENST00000351487.5 |
UPF3A
|
UPF3A regulator of nonsense mediated mRNA decay |
| chr3_-_134029914 | 0.01 |
ENST00000493729.5
ENST00000310926.11 |
SLCO2A1
|
solute carrier organic anion transporter family member 2A1 |
| chr4_-_145938775 | 0.01 |
ENST00000508784.6
|
ZNF827
|
zinc finger protein 827 |
| chr9_-_114930508 | 0.01 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr1_+_26787667 | 0.01 |
ENST00000674335.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr15_-_82806054 | 0.01 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr11_-_74949079 | 0.01 |
ENST00000528219.5
ENST00000684022.1 ENST00000531852.5 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr4_-_145938422 | 0.01 |
ENST00000656985.1
ENST00000652097.1 ENST00000503462.3 ENST00000379448.9 ENST00000513840.2 |
ZNF827
|
zinc finger protein 827 |
| chr11_-_74949142 | 0.01 |
ENST00000321448.12
ENST00000340360.10 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr11_-_74949114 | 0.01 |
ENST00000527087.5
|
XRRA1
|
X-ray radiation resistance associated 1 |
| chr5_-_60945008 | 0.01 |
ENST00000683052.1
ENST00000675229.1 ENST00000265038.10 ENST00000676185.1 ENST00000682217.1 ENST00000439176.6 ENST00000675042.2 |
ERCC8
|
ERCC excision repair 8, CSA ubiquitin ligase complex subunit |
| chr11_+_74949241 | 0.01 |
ENST00000610881.4
ENST00000530257.5 ENST00000526361.1 ENST00000532972.5 ENST00000263672.11 |
SPCS2
|
signal peptidase complex subunit 2 |
| chr12_-_110742929 | 0.00 |
ENST00000340766.9
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr10_-_101229449 | 0.00 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr11_-_119101814 | 0.00 |
ENST00000682791.1
ENST00000639704.1 ENST00000354202.9 |
DPAGT1
|
dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| chr14_-_65102468 | 0.00 |
ENST00000555932.5
ENST00000284165.10 ENST00000358402.8 ENST00000246163.2 ENST00000358664.9 ENST00000556979.5 ENST00000555667.5 ENST00000557746.5 ENST00000556443.5 ENST00000618858.4 ENST00000557277.5 ENST00000556892.5 |
MAX
|
MYC associated factor X |
| chr3_+_142596385 | 0.00 |
ENST00000457734.7
ENST00000483373.5 ENST00000475296.5 ENST00000495744.5 ENST00000476044.5 ENST00000461644.5 ENST00000464320.5 |
PLS1
|
plastin 1 |
| chr17_-_41047267 | 0.00 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr12_-_110742839 | 0.00 |
ENST00000551676.5
ENST00000550991.5 ENST00000335007.10 |
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr14_+_90398159 | 0.00 |
ENST00000544280.6
|
CALM1
|
calmodulin 1 |
| chrX_+_123961696 | 0.00 |
ENST00000371145.8
ENST00000371157.7 ENST00000371144.7 |
STAG2
|
stromal antigen 2 |
| chr7_-_28180735 | 0.00 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr1_-_24930263 | 0.00 |
ENST00000308873.11
|
RUNX3
|
RUNX family transcription factor 3 |
| chr2_+_157257687 | 0.00 |
ENST00000259056.5
|
GALNT5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_+_11556337 | 0.00 |
ENST00000234142.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr17_+_27471999 | 0.00 |
ENST00000583370.5
ENST00000509603.6 ENST00000268763.10 ENST00000398988.7 |
KSR1
|
kinase suppressor of ras 1 |
| chr4_+_87650277 | 0.00 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr20_+_2536573 | 0.00 |
ENST00000358864.2
|
TMC2
|
transmembrane channel like 2 |
| chrM_+_10055 | 0.00 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.3 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061598 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |