Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for TEAD4
Z-value: 0.51
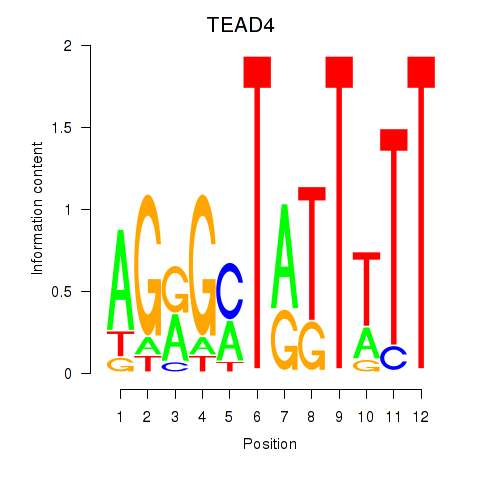
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.10 | TEAD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg38_v1_chr12_+_2959296_2959431, hg38_v1_chr12_+_2959870_2959953 | -0.03 | 8.8e-01 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_227813834 | 1.18 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr4_-_109801978 | 0.69 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr12_-_91146195 | 0.69 |
ENST00000548218.1
|
DCN
|
decorin |
| chr1_+_20589044 | 0.60 |
ENST00000375071.4
|
CDA
|
cytidine deaminase |
| chr6_-_132659178 | 0.59 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr7_-_41703062 | 0.50 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr2_-_234497035 | 0.48 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr11_-_102530738 | 0.47 |
ENST00000260227.5
|
MMP7
|
matrix metallopeptidase 7 |
| chr19_+_35139440 | 0.45 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_160200289 | 0.45 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr7_+_18495723 | 0.43 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr8_-_27258414 | 0.42 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr12_-_91178520 | 0.42 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr9_+_72577369 | 0.41 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr19_+_35139724 | 0.41 |
ENST00000588715.5
ENST00000588607.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_160200251 | 0.40 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr2_+_190927649 | 0.40 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr6_-_159726871 | 0.39 |
ENST00000535561.5
|
SOD2
|
superoxide dismutase 2 |
| chr2_-_160200310 | 0.38 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr19_+_35138993 | 0.35 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr5_+_174045673 | 0.34 |
ENST00000303177.8
ENST00000519867.5 |
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr4_-_144140683 | 0.34 |
ENST00000324022.14
|
GYPA
|
glycophorin A (MNS blood group) |
| chr7_+_80638633 | 0.31 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr2_-_210303608 | 0.30 |
ENST00000341685.8
|
MYL1
|
myosin light chain 1 |
| chr6_-_76072654 | 0.30 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr11_-_128587551 | 0.30 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr1_+_89633086 | 0.29 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C |
| chr18_+_49562049 | 0.28 |
ENST00000261292.9
ENST00000427224.6 ENST00000580036.5 |
LIPG
|
lipase G, endothelial type |
| chr4_-_144140635 | 0.28 |
ENST00000512064.5
ENST00000512789.5 ENST00000504786.5 ENST00000503627.2 ENST00000642713.1 ENST00000643148.1 ENST00000642738.1 ENST00000642295.1 ENST00000646447.1 ENST00000535709.6 |
GYPA
|
glycophorin A (MNS blood group) |
| chr2_-_127220293 | 0.27 |
ENST00000664447.2
ENST00000409327.2 |
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1 |
| chr10_-_73651013 | 0.26 |
ENST00000372873.8
|
SYNPO2L
|
synaptopodin 2 like |
| chr2_-_232776555 | 0.26 |
ENST00000438786.1
ENST00000233826.4 ENST00000409779.1 |
KCNJ13
|
potassium inwardly rectifying channel subfamily J member 13 |
| chr17_-_40984297 | 0.26 |
ENST00000377755.9
|
KRT40
|
keratin 40 |
| chr8_-_27258386 | 0.25 |
ENST00000350889.8
ENST00000519997.5 ENST00000519614.5 ENST00000522908.1 ENST00000265770.11 |
STMN4
|
stathmin 4 |
| chr11_-_10568571 | 0.25 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr20_+_61599755 | 0.25 |
ENST00000543233.2
|
CDH4
|
cadherin 4 |
| chr3_+_124094663 | 0.24 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr18_-_70205655 | 0.23 |
ENST00000255674.11
ENST00000640769.2 |
RTTN
|
rotatin |
| chr15_-_19988117 | 0.23 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr14_-_106803221 | 0.23 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr12_+_32107296 | 0.22 |
ENST00000551086.1
|
BICD1
|
BICD cargo adaptor 1 |
| chr12_+_10307950 | 0.22 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr3_+_112086335 | 0.22 |
ENST00000431717.6
ENST00000480282.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr6_-_46015812 | 0.22 |
ENST00000544153.3
ENST00000339561.12 |
CLIC5
|
chloride intracellular channel 5 |
| chr6_-_25830557 | 0.21 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr11_-_118925916 | 0.21 |
ENST00000683865.1
|
BCL9L
|
BCL9 like |
| chr8_+_22367526 | 0.21 |
ENST00000289952.9
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 member 14 |
| chr20_+_1266263 | 0.21 |
ENST00000649598.1
ENST00000381867.6 ENST00000381873.7 |
SNPH
|
syntaphilin |
| chr3_-_185821092 | 0.21 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr14_-_106360320 | 0.21 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr6_-_29431967 | 0.21 |
ENST00000377154.1
ENST00000641152.2 |
OR5V1
OR11A1
|
olfactory receptor family 5 subfamily V member 1 olfactory receptor family 11 subfamily A member 1 |
| chr10_-_88952763 | 0.20 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr18_-_3219961 | 0.20 |
ENST00000356443.9
|
MYOM1
|
myomesin 1 |
| chr7_+_77538027 | 0.19 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr4_+_41612702 | 0.19 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612892 | 0.19 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_40750302 | 0.19 |
ENST00000598485.6
ENST00000378313.7 ENST00000470681.5 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr2_+_33476640 | 0.18 |
ENST00000425210.5
ENST00000444784.5 ENST00000423159.5 ENST00000403687.8 |
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr4_-_26490453 | 0.18 |
ENST00000295589.4
|
CCKAR
|
cholecystokinin A receptor |
| chr22_-_50085414 | 0.18 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr7_-_47948448 | 0.18 |
ENST00000289672.7
|
PKD1L1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr14_+_78403686 | 0.17 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr10_+_122163672 | 0.17 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr1_-_156282799 | 0.17 |
ENST00000361813.5
|
SMG5
|
SMG5 nonsense mediated mRNA decay factor |
| chr22_-_50085331 | 0.17 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr10_-_13234368 | 0.16 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr11_-_10568650 | 0.16 |
ENST00000256178.8
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_+_66493556 | 0.16 |
ENST00000305454.8
ENST00000522977.5 ENST00000480005.1 |
VXN
|
vexin |
| chrX_-_41665766 | 0.16 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr10_-_13234329 | 0.16 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chr11_+_6845683 | 0.16 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr7_+_151114597 | 0.16 |
ENST00000335367.7
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_-_211492111 | 0.16 |
ENST00000367002.5
ENST00000680073.1 |
RD3
|
RD3 regulator of GUCY2D |
| chr3_-_112638097 | 0.16 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr3_-_33645433 | 0.16 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_+_119135825 | 0.16 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr1_+_15684284 | 0.15 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr7_+_77538059 | 0.15 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr6_-_13328332 | 0.15 |
ENST00000606530.1
ENST00000607658.5 ENST00000356436.8 ENST00000450347.5 ENST00000452989.5 ENST00000422136.5 ENST00000446018.5 ENST00000379291.5 ENST00000379300.8 ENST00000606370.5 ENST00000343141.8 ENST00000379307.6 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family member 7 |
| chr11_-_55936400 | 0.15 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr2_-_187554351 | 0.15 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr12_+_56041893 | 0.14 |
ENST00000552361.1
ENST00000646449.2 |
RPS26
|
ribosomal protein S26 |
| chr15_+_81182579 | 0.14 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr1_-_151826085 | 0.14 |
ENST00000356728.11
|
RORC
|
RAR related orphan receptor C |
| chr9_-_127847117 | 0.14 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr12_+_10307818 | 0.14 |
ENST00000350274.9
ENST00000336164.9 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr14_-_94517844 | 0.13 |
ENST00000341228.2
|
SERPINA12
|
serpin family A member 12 |
| chr2_-_162318129 | 0.13 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr18_+_58195390 | 0.13 |
ENST00000456173.6
ENST00000676226.1 ENST00000675865.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr15_+_80441229 | 0.13 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr14_-_106593319 | 0.13 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr20_-_13990609 | 0.13 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr17_-_28365244 | 0.13 |
ENST00000536498.5
|
SEBOX
|
SEBOX homeobox |
| chr19_+_9087061 | 0.13 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr11_+_62242232 | 0.12 |
ENST00000244926.4
|
SCGB1D2
|
secretoglobin family 1D member 2 |
| chr5_-_139482714 | 0.12 |
ENST00000652543.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr12_+_68808143 | 0.12 |
ENST00000258149.11
ENST00000428863.6 ENST00000393412.7 |
MDM2
|
MDM2 proto-oncogene |
| chr1_+_178341445 | 0.12 |
ENST00000462775.5
|
RASAL2
|
RAS protein activator like 2 |
| chr6_-_27867581 | 0.12 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr12_-_24903014 | 0.12 |
ENST00000539282.5
|
BCAT1
|
branched chain amino acid transaminase 1 |
| chr2_-_136118142 | 0.12 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr9_+_74497308 | 0.12 |
ENST00000376896.8
|
RORB
|
RAR related orphan receptor B |
| chr5_+_58583068 | 0.12 |
ENST00000282878.6
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr7_-_36985060 | 0.12 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_+_63443306 | 0.12 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr10_+_84452208 | 0.12 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr14_-_106335613 | 0.12 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr4_+_185395979 | 0.11 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr3_+_138348823 | 0.11 |
ENST00000475711.5
ENST00000464896.5 |
MRAS
|
muscle RAS oncogene homolog |
| chrX_+_37780049 | 0.11 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr21_-_38498415 | 0.11 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr5_+_93583212 | 0.11 |
ENST00000327111.8
|
NR2F1
|
nuclear receptor subfamily 2 group F member 1 |
| chr3_+_63443076 | 0.11 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr6_-_32666648 | 0.11 |
ENST00000399082.7
ENST00000399079.7 ENST00000374943.8 ENST00000434651.6 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr4_-_154590735 | 0.11 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr1_-_101996919 | 0.11 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr18_+_34710307 | 0.10 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha |
| chr4_-_142305935 | 0.10 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_-_39032343 | 0.10 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr2_-_150487658 | 0.10 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr4_-_88231322 | 0.10 |
ENST00000515655.5
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr3_+_149474688 | 0.10 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_+_9185594 | 0.10 |
ENST00000344248.4
|
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr15_+_59438149 | 0.10 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr20_-_56525925 | 0.10 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chrX_+_80420466 | 0.10 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr3_+_124094696 | 0.09 |
ENST00000360013.7
ENST00000684186.1 ENST00000684276.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr5_+_54455661 | 0.09 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3 |
| chr22_+_21420636 | 0.09 |
ENST00000407598.2
|
HIC2
|
HIC ZBTB transcriptional repressor 2 |
| chr5_-_139439488 | 0.09 |
ENST00000302060.10
|
DNAJC18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr18_-_3219849 | 0.09 |
ENST00000261606.11
|
MYOM1
|
myomesin 1 |
| chr12_+_12725897 | 0.09 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr5_-_95081482 | 0.09 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr7_-_108240049 | 0.09 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr18_+_58341038 | 0.09 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr4_-_47981535 | 0.09 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr9_+_68356603 | 0.09 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5 |
| chr9_+_113594118 | 0.09 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chr5_+_141330494 | 0.09 |
ENST00000517417.3
ENST00000378105.4 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr12_+_18262730 | 0.09 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr2_-_231125032 | 0.09 |
ENST00000258400.4
|
HTR2B
|
5-hydroxytryptamine receptor 2B |
| chr17_-_68955332 | 0.09 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr3_+_138348570 | 0.08 |
ENST00000423968.7
|
MRAS
|
muscle RAS oncogene homolog |
| chrX_+_21374288 | 0.08 |
ENST00000642359.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr8_+_49911604 | 0.08 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr7_+_54542362 | 0.08 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr6_-_39725335 | 0.08 |
ENST00000538893.5
|
KIF6
|
kinesin family member 6 |
| chr7_+_80638510 | 0.08 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr19_-_3786254 | 0.08 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr5_-_139482285 | 0.08 |
ENST00000652110.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr11_+_46936710 | 0.08 |
ENST00000378618.6
ENST00000278460.12 ENST00000395460.6 ENST00000378615.7 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr1_+_160081529 | 0.08 |
ENST00000368088.4
|
KCNJ9
|
potassium inwardly rectifying channel subfamily J member 9 |
| chr3_+_101827982 | 0.08 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr4_+_70430487 | 0.08 |
ENST00000413702.5
|
MUC7
|
mucin 7, secreted |
| chr17_-_41149823 | 0.08 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr6_+_31670167 | 0.08 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr14_+_21797272 | 0.08 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr14_-_36582593 | 0.07 |
ENST00000258829.6
|
NKX2-8
|
NK2 homeobox 8 |
| chr7_+_80638662 | 0.07 |
ENST00000394788.7
|
CD36
|
CD36 molecule |
| chr3_+_14675128 | 0.07 |
ENST00000435614.5
ENST00000253697.8 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr4_-_142305826 | 0.07 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr17_+_42458844 | 0.07 |
ENST00000393829.6
ENST00000537728.5 ENST00000343619.9 ENST00000264649.10 ENST00000585525.5 ENST00000544137.5 ENST00000589727.5 ENST00000587824.5 |
ATP6V0A1
|
ATPase H+ transporting V0 subunit a1 |
| chr8_+_49911396 | 0.07 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr15_-_65106308 | 0.07 |
ENST00000502113.2
|
UBAP1L
|
ubiquitin associated protein 1 like |
| chr4_+_54229261 | 0.07 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chr9_-_128191878 | 0.07 |
ENST00000538431.5
|
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr19_-_51417619 | 0.07 |
ENST00000441969.7
ENST00000339313.10 ENST00000525998.5 ENST00000436984.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr5_+_180988840 | 0.07 |
ENST00000342868.7
|
BTNL3
|
butyrophilin like 3 |
| chr18_-_13726510 | 0.07 |
ENST00000651643.1
ENST00000322247.7 |
FAM210A
|
family with sequence similarity 210 member A |
| chr1_+_65992389 | 0.07 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr7_+_148133684 | 0.07 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr2_+_209771972 | 0.06 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr19_-_3786408 | 0.06 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chrX_+_21374608 | 0.06 |
ENST00000644295.1
ENST00000645074.1 ENST00000645791.1 ENST00000643220.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr13_-_37869723 | 0.06 |
ENST00000426868.6
ENST00000379705.8 ENST00000338947.9 ENST00000355779.6 ENST00000358477.6 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel subfamily C member 4 |
| chr3_+_138348592 | 0.06 |
ENST00000621127.4
ENST00000494949.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr17_+_34255274 | 0.06 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr6_-_152168349 | 0.06 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr15_+_33968484 | 0.06 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr12_+_32107151 | 0.06 |
ENST00000548411.5
|
BICD1
|
BICD cargo adaptor 1 |
| chr19_-_3786363 | 0.06 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr5_-_42811884 | 0.06 |
ENST00000514985.6
ENST00000511224.5 ENST00000507920.5 ENST00000510965.1 |
SELENOP
|
selenoprotein P |
| chr6_-_87702221 | 0.06 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chr6_-_152168291 | 0.06 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr11_-_84273188 | 0.06 |
ENST00000330014.11
ENST00000418306.6 ENST00000531015.5 |
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr12_-_9733083 | 0.06 |
ENST00000542530.5
|
CLECL1
|
C-type lectin like 1 |
| chr12_+_6444932 | 0.06 |
ENST00000266557.4
|
CD27
|
CD27 molecule |
| chr12_+_10179006 | 0.06 |
ENST00000298530.7
|
TMEM52B
|
transmembrane protein 52B |
| chr16_-_29862890 | 0.06 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr6_+_159726998 | 0.06 |
ENST00000614346.4
|
WTAP
|
WT1 associated protein |
| chrX_+_21374434 | 0.06 |
ENST00000279451.9
ENST00000645245.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr3_+_130560334 | 0.05 |
ENST00000358511.10
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr7_+_54542300 | 0.05 |
ENST00000302287.7
ENST00000407838.7 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr7_+_144003929 | 0.05 |
ENST00000408922.3
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr5_+_154712824 | 0.05 |
ENST00000336314.9
|
LARP1
|
La ribonucleoprotein 1, translational regulator |
| chr8_+_54457927 | 0.05 |
ENST00000297316.5
|
SOX17
|
SRY-box transcription factor 17 |
| chr22_-_30246739 | 0.05 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr14_-_106165730 | 0.05 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr7_+_23299306 | 0.05 |
ENST00000466681.2
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr1_-_179877790 | 0.05 |
ENST00000495650.1
ENST00000367612.7 ENST00000482587.5 ENST00000609928.6 |
TOR1AIP2
|
torsin 1A interacting protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 0.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.3 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:2000332 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.3 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.3 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.0 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.5 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.5 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 1.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.3 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |