Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
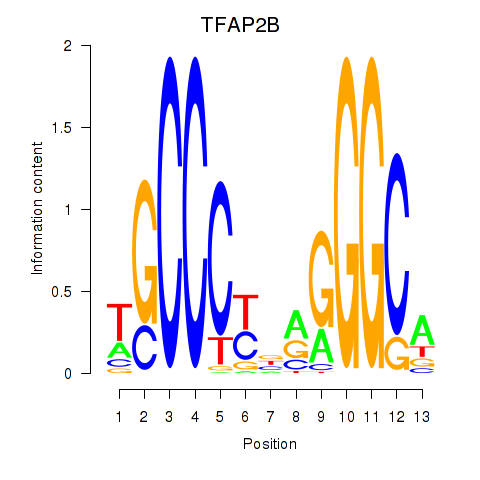
Results for TFAP2B
Z-value: 0.69
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.13 | TFAP2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2B | hg38_v1_chr6_+_50818701_50818723 | 0.20 | 2.8e-01 | Click! |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_149141817 | 1.58 |
ENST00000504238.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr5_+_149141483 | 1.51 |
ENST00000326685.11
ENST00000309868.12 |
ABLIM3
|
actin binding LIM protein family member 3 |
| chr5_+_149141573 | 1.48 |
ENST00000506113.5
|
ABLIM3
|
actin binding LIM protein family member 3 |
| chr12_-_89352395 | 1.13 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_89352487 | 1.03 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_152908538 | 0.96 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr11_-_61581104 | 0.95 |
ENST00000263846.8
|
SYT7
|
synaptotagmin 7 |
| chr19_-_11578937 | 0.93 |
ENST00000592659.1
ENST00000592828.6 ENST00000218758.9 ENST00000412435.6 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr14_+_94174284 | 0.85 |
ENST00000304338.8
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr9_+_87498491 | 0.73 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr21_-_26845402 | 0.70 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr1_+_17249088 | 0.70 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase 3 |
| chr14_+_64704380 | 0.66 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr17_-_5234801 | 0.64 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr1_-_20486197 | 0.64 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr1_-_74732997 | 0.62 |
ENST00000370872.7
ENST00000370871.7 ENST00000340866.10 ENST00000370870.5 |
CRYZ
|
crystallin zeta |
| chr1_-_74733253 | 0.60 |
ENST00000417775.5
|
CRYZ
|
crystallin zeta |
| chr9_+_87497852 | 0.57 |
ENST00000408954.8
|
DAPK1
|
death associated protein kinase 1 |
| chr19_+_35154715 | 0.56 |
ENST00000392218.6
ENST00000543307.5 ENST00000392219.7 ENST00000541435.6 ENST00000590686.5 ENST00000342879.7 ENST00000588699.5 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr2_+_95346649 | 0.56 |
ENST00000468529.1
|
KCNIP3
|
potassium voltage-gated channel interacting protein 3 |
| chr20_+_63696643 | 0.56 |
ENST00000369996.3
|
TNFRSF6B
|
TNF receptor superfamily member 6b |
| chr8_-_143572748 | 0.56 |
ENST00000529971.1
ENST00000398882.8 |
MROH6
|
maestro heat like repeat family member 6 |
| chr19_+_35154914 | 0.56 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr11_-_68213828 | 0.54 |
ENST00000405515.5
|
KMT5B
|
lysine methyltransferase 5B |
| chr1_+_155208727 | 0.54 |
ENST00000316721.8
|
MTX1
|
metaxin 1 |
| chr19_+_50203607 | 0.54 |
ENST00000642316.2
ENST00000425460.6 ENST00000440075.6 ENST00000376970.6 ENST00000599920.5 |
MYH14
|
myosin heavy chain 14 |
| chr9_+_17579059 | 0.54 |
ENST00000380607.5
|
SH3GL2
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr1_+_7784411 | 0.53 |
ENST00000613533.4
ENST00000614998.4 |
PER3
|
period circadian regulator 3 |
| chr17_+_45620323 | 0.53 |
ENST00000634540.1
|
LINC02210-CRHR1
|
LINC02210-CRHR1 readthrough |
| chr14_+_54567100 | 0.51 |
ENST00000554335.6
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr18_+_3449413 | 0.51 |
ENST00000549253.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr22_+_31082860 | 0.49 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr1_+_155033824 | 0.49 |
ENST00000295542.6
ENST00000423025.6 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr2_-_207165923 | 0.49 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr1_+_155208690 | 0.49 |
ENST00000368376.8
|
MTX1
|
metaxin 1 |
| chr14_+_79279906 | 0.49 |
ENST00000428277.6
|
NRXN3
|
neurexin 3 |
| chr14_+_79279403 | 0.48 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr14_+_94174334 | 0.48 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4 regulatory subunit 4 |
| chr11_+_126355634 | 0.48 |
ENST00000227495.10
ENST00000676545.1 ENST00000678865.1 ENST00000444328.7 ENST00000677503.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr11_-_68213577 | 0.46 |
ENST00000402789.5
ENST00000402185.6 ENST00000458496.1 |
KMT5B
|
lysine methyltransferase 5B |
| chr15_-_79090760 | 0.44 |
ENST00000419573.7
ENST00000558480.7 |
RASGRF1
|
Ras protein specific guanine nucleotide releasing factor 1 |
| chr11_-_7673485 | 0.44 |
ENST00000299498.11
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr16_+_30949054 | 0.44 |
ENST00000318663.5
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr3_-_197029775 | 0.44 |
ENST00000439320.1
ENST00000296351.8 ENST00000296350.10 |
MELTF
|
melanotransferrin |
| chr4_+_94489030 | 0.43 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr1_+_155209213 | 0.43 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr11_-_66336396 | 0.42 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr14_+_93927358 | 0.42 |
ENST00000557000.2
|
FAM181A
|
family with sequence similarity 181 member A |
| chr11_+_126355894 | 0.41 |
ENST00000530591.5
ENST00000534083.5 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr5_+_170353480 | 0.41 |
ENST00000377360.8
|
KCNIP1
|
potassium voltage-gated channel interacting protein 1 |
| chr19_+_1067144 | 0.41 |
ENST00000313093.7
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr19_+_1067272 | 0.41 |
ENST00000590214.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr19_+_1067493 | 0.40 |
ENST00000586866.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr16_+_30896606 | 0.40 |
ENST00000279804.3
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chr11_-_7674206 | 0.39 |
ENST00000533558.5
ENST00000527542.5 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr4_+_1721470 | 0.39 |
ENST00000612220.5
ENST00000313288.9 |
TACC3
|
transforming acidic coiled-coil containing protein 3 |
| chr15_+_58771280 | 0.37 |
ENST00000559228.6
ENST00000450403.3 |
MINDY2
|
MINDY lysine 48 deubiquitinase 2 |
| chr3_-_52231190 | 0.37 |
ENST00000494383.1
|
ENSG00000173366.11
|
novel twinfilin, actin-binding protein, homolog 2 (Drosophila) (TWF2) and toll-like receptor 9 (TLR9) protein |
| chr21_+_42219123 | 0.37 |
ENST00000398449.8
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr20_+_6767678 | 0.37 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr9_+_121651594 | 0.36 |
ENST00000408936.7
|
DAB2IP
|
DAB2 interacting protein |
| chr1_+_31576485 | 0.36 |
ENST00000457433.6
ENST00000271064.12 |
TINAGL1
|
tubulointerstitial nephritis antigen like 1 |
| chr10_+_69318831 | 0.35 |
ENST00000359426.7
|
HK1
|
hexokinase 1 |
| chr11_+_61228377 | 0.35 |
ENST00000537932.5
|
PGA4
|
pepsinogen A4 |
| chr21_+_42219111 | 0.35 |
ENST00000450121.5
ENST00000361802.6 |
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr6_-_30686624 | 0.35 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr19_+_10718114 | 0.35 |
ENST00000408974.8
|
DNM2
|
dynamin 2 |
| chr7_-_100468063 | 0.35 |
ENST00000423266.5
ENST00000456330.1 |
TSC22D4
|
TSC22 domain family member 4 |
| chr1_-_25906457 | 0.33 |
ENST00000426559.6
|
STMN1
|
stathmin 1 |
| chr2_+_200440649 | 0.33 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chrX_+_7147675 | 0.33 |
ENST00000674429.1
|
STS
|
steroid sulfatase |
| chrX_-_46759055 | 0.33 |
ENST00000328306.4
ENST00000616978.5 |
SLC9A7
|
solute carrier family 9 member A7 |
| chr10_-_98030612 | 0.33 |
ENST00000370597.8
|
CRTAC1
|
cartilage acidic protein 1 |
| chr1_+_11934651 | 0.33 |
ENST00000449038.5
ENST00000196061.5 ENST00000429000.6 |
PLOD1
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 |
| chr15_-_74209019 | 0.33 |
ENST00000323940.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr2_-_219309484 | 0.33 |
ENST00000409251.7
ENST00000451506.5 ENST00000446182.5 |
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr19_+_40601342 | 0.32 |
ENST00000396819.8
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_-_45425620 | 0.32 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr16_+_2964216 | 0.32 |
ENST00000572045.5
ENST00000571007.5 ENST00000575885.5 ENST00000303746.10 ENST00000319500.10 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr10_-_70170466 | 0.32 |
ENST00000373239.2
ENST00000373241.9 ENST00000373242.6 |
SAR1A
|
secretion associated Ras related GTPase 1A |
| chr6_-_30687200 | 0.32 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr10_+_133527355 | 0.31 |
ENST00000252945.8
ENST00000421586.5 ENST00000418356.1 |
CYP2E1
|
cytochrome P450 family 2 subfamily E member 1 |
| chr1_-_237004440 | 0.31 |
ENST00000464121.3
|
MT1HL1
|
metallothionein 1H like 1 |
| chr13_+_20703677 | 0.31 |
ENST00000682841.1
|
IL17D
|
interleukin 17D |
| chr9_-_120877026 | 0.31 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19 |
| chr1_+_206507546 | 0.31 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr16_-_636253 | 0.30 |
ENST00000565163.5
|
METTL26
|
methyltransferase like 26 |
| chr17_-_43778937 | 0.30 |
ENST00000226004.8
|
DUSP3
|
dual specificity phosphatase 3 |
| chrX_+_7147237 | 0.29 |
ENST00000666110.2
|
STS
|
steroid sulfatase |
| chr2_-_227164194 | 0.29 |
ENST00000396625.5
|
COL4A4
|
collagen type IV alpha 4 chain |
| chr15_-_25865076 | 0.29 |
ENST00000619904.1
|
ATP10A
|
ATPase phospholipid transporting 10A (putative) |
| chr12_+_112418889 | 0.29 |
ENST00000392597.5
|
PTPN11
|
protein tyrosine phosphatase non-receptor type 11 |
| chr9_+_128420812 | 0.29 |
ENST00000372838.9
|
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr11_+_73289403 | 0.29 |
ENST00000535931.2
ENST00000544437.6 |
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr9_+_131289685 | 0.29 |
ENST00000372264.4
|
PLPP7
|
phospholipid phosphatase 7 (inactive) |
| chr2_-_219309350 | 0.29 |
ENST00000295718.7
|
PTPRN
|
protein tyrosine phosphatase receptor type N |
| chr8_-_142786530 | 0.29 |
ENST00000301263.5
|
LY6D
|
lymphocyte antigen 6 family member D |
| chr1_+_206507520 | 0.28 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr20_-_46308485 | 0.28 |
ENST00000537909.4
|
CDH22
|
cadherin 22 |
| chr8_-_141001217 | 0.28 |
ENST00000522684.5
ENST00000524357.5 ENST00000521332.5 ENST00000524040.5 ENST00000519881.5 ENST00000520045.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr8_-_143973520 | 0.28 |
ENST00000356346.7
|
PLEC
|
plectin |
| chr6_+_33410961 | 0.28 |
ENST00000374512.7
ENST00000374516.8 |
PHF1
|
PHD finger protein 1 |
| chr2_-_27263034 | 0.28 |
ENST00000233535.9
|
SLC30A3
|
solute carrier family 30 member 3 |
| chr11_-_61580826 | 0.28 |
ENST00000540677.5
ENST00000542836.5 ENST00000542670.5 ENST00000535826.5 ENST00000545053.1 ENST00000539008.6 |
SYT7
|
synaptotagmin 7 |
| chrX_+_132023580 | 0.27 |
ENST00000496850.1
|
STK26
|
serine/threonine kinase 26 |
| chr5_+_150601060 | 0.27 |
ENST00000394243.5
|
SYNPO
|
synaptopodin |
| chr11_+_43942627 | 0.27 |
ENST00000617612.3
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr5_-_177303675 | 0.27 |
ENST00000393611.6
ENST00000303270.6 ENST00000303251.11 |
RAB24
|
RAB24, member RAS oncogene family |
| chr13_-_19863508 | 0.27 |
ENST00000382907.8
ENST00000382905.8 |
ZMYM5
|
zinc finger MYM-type containing 5 |
| chr12_-_52796110 | 0.27 |
ENST00000417996.2
|
KRT3
|
keratin 3 |
| chr16_-_636270 | 0.27 |
ENST00000397665.6
ENST00000397666.6 ENST00000301686.13 ENST00000338401.8 ENST00000397664.8 ENST00000568830.1 ENST00000614890.4 |
METTL26
|
methyltransferase like 26 |
| chr22_+_18529042 | 0.26 |
ENST00000613577.4
|
TMEM191B
|
transmembrane protein 191B |
| chr19_-_42242526 | 0.26 |
ENST00000222330.8
ENST00000676537.1 |
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr1_-_204213943 | 0.26 |
ENST00000308302.4
|
GOLT1A
|
golgi transport 1A |
| chr21_+_43728851 | 0.26 |
ENST00000327574.4
|
PDXK
|
pyridoxal kinase |
| chr13_-_21459226 | 0.26 |
ENST00000320220.13
|
ZDHHC20
|
zinc finger DHHC-type palmitoyltransferase 20 |
| chr19_-_1822038 | 0.26 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog |
| chr16_-_71289609 | 0.25 |
ENST00000338099.9
ENST00000563876.1 |
CMTR2
|
cap methyltransferase 2 |
| chr17_-_48037760 | 0.25 |
ENST00000621465.5
|
COPZ2
|
COPI coat complex subunit zeta 2 |
| chr14_+_79279339 | 0.25 |
ENST00000557594.5
|
NRXN3
|
neurexin 3 |
| chr14_+_79279681 | 0.25 |
ENST00000679122.1
|
NRXN3
|
neurexin 3 |
| chr14_-_91253925 | 0.25 |
ENST00000531499.2
|
GPR68
|
G protein-coupled receptor 68 |
| chr1_+_178725147 | 0.25 |
ENST00000367634.6
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr19_-_55149193 | 0.25 |
ENST00000587758.5
ENST00000588981.6 ENST00000356783.9 ENST00000291901.12 ENST00000588426.5 ENST00000536926.5 ENST00000588147.5 |
TNNT1
|
troponin T1, slow skeletal type |
| chr10_+_11005301 | 0.25 |
ENST00000416382.6
ENST00000631460.1 ENST00000631816.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr3_+_100492548 | 0.25 |
ENST00000323523.8
ENST00000403410.5 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr9_-_34522968 | 0.24 |
ENST00000399775.3
|
ENHO
|
energy homeostasis associated |
| chr12_+_131894615 | 0.24 |
ENST00000321867.6
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr5_+_172641241 | 0.24 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr1_-_27012837 | 0.24 |
ENST00000289166.6
|
TENT5B
|
terminal nucleotidyltransferase 5B |
| chr5_+_150190035 | 0.24 |
ENST00000230671.7
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 member 7 |
| chr17_-_80476597 | 0.24 |
ENST00000306773.5
|
NPTX1
|
neuronal pentraxin 1 |
| chr17_-_5584448 | 0.24 |
ENST00000269280.8
ENST00000571451.6 ENST00000572272.6 ENST00000613500.4 ENST00000619223.4 ENST00000617618.4 ENST00000345221.7 ENST00000262467.10 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr19_+_45467988 | 0.24 |
ENST00000615753.4
ENST00000585836.5 ENST00000417353.6 ENST00000591858.5 ENST00000443841.6 ENST00000590335.1 ENST00000353609.8 |
FOSB
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr14_-_93115812 | 0.24 |
ENST00000553452.5
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chrX_+_49058966 | 0.24 |
ENST00000496529.6
ENST00000606812.5 ENST00000603986.6 ENST00000536628.3 |
CCDC120
|
coiled-coil domain containing 120 |
| chr8_-_143944737 | 0.24 |
ENST00000398774.6
|
PLEC
|
plectin |
| chr10_+_18400562 | 0.23 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr1_+_178725227 | 0.23 |
ENST00000367635.8
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr19_+_41443130 | 0.23 |
ENST00000378187.3
|
ERICH4
|
glutamate rich 4 |
| chr22_-_18024513 | 0.23 |
ENST00000441493.7
|
MICAL3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chrX_+_69504320 | 0.23 |
ENST00000252338.5
|
FAM155B
|
family with sequence similarity 155 member B |
| chr16_+_2817230 | 0.23 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr16_-_71289367 | 0.23 |
ENST00000434935.7
ENST00000565850.1 ENST00000568910.1 |
CMTR2
|
cap methyltransferase 2 |
| chr2_+_184598520 | 0.23 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chr9_-_120877167 | 0.23 |
ENST00000373896.8
ENST00000312189.10 |
PHF19
|
PHD finger protein 19 |
| chr1_+_45326869 | 0.23 |
ENST00000334815.6
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase like |
| chr20_+_46118277 | 0.23 |
ENST00000620709.4
|
CD40
|
CD40 molecule |
| chr5_-_135034212 | 0.23 |
ENST00000265340.12
|
PITX1
|
paired like homeodomain 1 |
| chr5_-_157575741 | 0.23 |
ENST00000517905.1
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr1_-_42740140 | 0.22 |
ENST00000372539.3
ENST00000296387.6 ENST00000539749.5 |
CLDN19
|
claudin 19 |
| chr22_-_38088915 | 0.22 |
ENST00000428572.1
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr1_-_25906411 | 0.22 |
ENST00000455785.7
|
STMN1
|
stathmin 1 |
| chr6_+_54846735 | 0.22 |
ENST00000306858.8
|
FAM83B
|
family with sequence similarity 83 member B |
| chr9_+_75890639 | 0.22 |
ENST00000545128.5
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr19_+_2360238 | 0.22 |
ENST00000649857.1
|
TMPRSS9
|
transmembrane serine protease 9 |
| chr12_+_109573757 | 0.22 |
ENST00000228510.8
ENST00000539696.5 ENST00000392727.7 |
MVK
|
mevalonate kinase |
| chr7_-_27130182 | 0.22 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr12_-_47758828 | 0.22 |
ENST00000389212.7
ENST00000449771.7 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr14_+_74348440 | 0.22 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr1_-_153615858 | 0.22 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr15_+_73684373 | 0.22 |
ENST00000558689.5
ENST00000560786.6 ENST00000318443.10 ENST00000561213.5 |
CD276
|
CD276 molecule |
| chr8_+_38901327 | 0.21 |
ENST00000519640.5
ENST00000617275.5 |
PLEKHA2
|
pleckstrin homology domain containing A2 |
| chr11_+_63839086 | 0.21 |
ENST00000350490.11
ENST00000402010.8 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr2_+_85133376 | 0.21 |
ENST00000282111.4
|
TCF7L1
|
transcription factor 7 like 1 |
| chr13_+_97434154 | 0.21 |
ENST00000245304.5
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr17_-_63700100 | 0.21 |
ENST00000578993.5
ENST00000259006.8 ENST00000583211.5 |
LIMD2
|
LIM domain containing 2 |
| chr22_-_28679865 | 0.21 |
ENST00000397906.6
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr18_+_58671439 | 0.21 |
ENST00000649217.2
|
MALT1
|
MALT1 paracaspase |
| chr16_-_11256192 | 0.21 |
ENST00000644787.1
ENST00000332029.4 |
SOCS1
|
suppressor of cytokine signaling 1 |
| chr11_+_63839173 | 0.21 |
ENST00000502399.7
ENST00000425897.3 ENST00000513765.7 ENST00000679216.1 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr21_+_44300038 | 0.20 |
ENST00000349048.9
ENST00000628044.1 |
PFKL
|
phosphofructokinase, liver type |
| chr19_-_19628197 | 0.20 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.8 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr14_+_90061979 | 0.20 |
ENST00000282146.5
|
KCNK13
|
potassium two pore domain channel subfamily K member 13 |
| chr9_-_128771909 | 0.20 |
ENST00000291900.7
ENST00000414921.5 |
ZER1
|
zyg-11 related cell cycle regulator |
| chr19_+_41797147 | 0.20 |
ENST00000596544.1
|
CEACAM3
|
CEA cell adhesion molecule 3 |
| chr2_-_240820945 | 0.19 |
ENST00000428768.2
ENST00000650053.1 ENST00000650130.1 |
KIF1A
|
kinesin family member 1A |
| chrX_+_49171889 | 0.19 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2 |
| chr17_+_7855055 | 0.19 |
ENST00000574668.1
ENST00000301599.7 |
TMEM88
|
transmembrane protein 88 |
| chrX_+_21940693 | 0.19 |
ENST00000404933.7
ENST00000379404.5 |
SMS
|
spermine synthase |
| chr9_-_113299196 | 0.19 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr11_-_64246907 | 0.19 |
ENST00000309318.8
|
PPP1R14B
|
protein phosphatase 1 regulatory inhibitor subunit 14B |
| chr7_-_143408848 | 0.19 |
ENST00000275815.4
|
EPHA1
|
EPH receptor A1 |
| chr16_+_67199509 | 0.19 |
ENST00000477898.5
|
ELMO3
|
engulfment and cell motility 3 |
| chr3_+_179148341 | 0.18 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr17_-_8190154 | 0.18 |
ENST00000389017.6
|
BORCS6
|
BLOC-1 related complex subunit 6 |
| chr9_-_98708856 | 0.18 |
ENST00000259455.4
|
GABBR2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr16_+_68644988 | 0.18 |
ENST00000429102.6
|
CDH3
|
cadherin 3 |
| chr16_-_636230 | 0.18 |
ENST00000568773.1
|
METTL26
|
methyltransferase like 26 |
| chr17_+_39667964 | 0.18 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr19_+_35031263 | 0.18 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr10_+_87863595 | 0.18 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog |
| chrX_-_19887585 | 0.18 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chrX_+_132023294 | 0.18 |
ENST00000481105.5
ENST00000354719.10 ENST00000394334.7 ENST00000394335.6 |
STK26
|
serine/threonine kinase 26 |
| chrX_-_153830527 | 0.18 |
ENST00000393758.7
ENST00000544474.5 |
PDZD4
|
PDZ domain containing 4 |
| chr13_+_96090098 | 0.18 |
ENST00000376705.4
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr1_-_97921042 | 0.18 |
ENST00000306031.5
|
DPYD
|
dihydropyrimidine dehydrogenase |
| chr10_-_97633485 | 0.18 |
ENST00000370635.3
ENST00000478953.1 ENST00000307450.11 |
MORN4
|
MORN repeat containing 4 |
| chr16_+_28824116 | 0.18 |
ENST00000568266.5
|
ATXN2L
|
ataxin 2 like |
| chr19_+_35704540 | 0.18 |
ENST00000392197.7
ENST00000426659.6 |
ZBTB32
|
zinc finger and BTB domain containing 32 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.4 | 1.2 | GO:1990927 | short-term synaptic potentiation(GO:1990926) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 1.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 | 1.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.7 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.2 | 0.5 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.2 | 0.6 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.2 | 0.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.7 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 0.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.4 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.1 | 1.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.4 | GO:0048817 | negative regulation of transforming growth factor beta2 production(GO:0032912) negative regulation of hair follicle maturation(GO:0048817) regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.3 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.6 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 1.4 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0051586 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.4 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.3 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.1 | 0.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.7 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0036018 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 1.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 1.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 4.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0097374 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 1.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.0 | 0.2 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.3 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.4 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.6 | GO:0071622 | regulation of granulocyte chemotaxis(GO:0071622) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.9 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.2 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 5.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.6 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 0.7 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.9 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 0.7 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.7 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.2 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 2.4 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.3 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 1.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.2 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.3 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |