Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
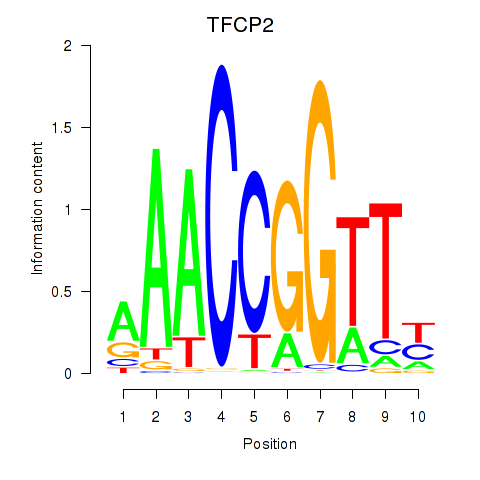
Results for TFCP2
Z-value: 0.92
Transcription factors associated with TFCP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2
|
ENSG00000135457.10 | TFCP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2 | hg38_v1_chr12_-_51173067_51173145 | -0.56 | 1.3e-03 | Click! |
Activity profile of TFCP2 motif
Sorted Z-values of TFCP2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_72577939 | 5.26 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr2_-_112784486 | 3.62 |
ENST00000263339.4
|
IL1A
|
interleukin 1 alpha |
| chr5_-_150289941 | 3.01 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr14_+_75280078 | 2.83 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_80073465 | 2.83 |
ENST00000404191.5
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr5_-_150289764 | 2.78 |
ENST00000671881.1
ENST00000672752.1 ENST00000510347.2 ENST00000672829.1 ENST00000348628.11 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr9_+_128420812 | 2.70 |
ENST00000372838.9
|
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr5_-_150290093 | 2.48 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr9_+_72577788 | 2.30 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr9_+_72577369 | 2.27 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr1_+_44746401 | 2.23 |
ENST00000372217.5
|
KIF2C
|
kinesin family member 2C |
| chr5_-_150289625 | 2.20 |
ENST00000683332.1
ENST00000398376.8 ENST00000672785.1 ENST00000672396.1 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr12_+_40692413 | 2.14 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr10_+_86958557 | 2.09 |
ENST00000372017.4
ENST00000348795.8 |
SNCG
|
synuclein gamma |
| chr15_-_74203172 | 2.06 |
ENST00000616000.4
|
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr10_-_88851809 | 2.05 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_+_62881686 | 1.74 |
ENST00000536981.6
ENST00000539891.6 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr20_-_5610980 | 1.70 |
ENST00000379019.7
|
GPCPD1
|
glycerophosphocholine phosphodiesterase 1 |
| chr5_+_138179093 | 1.68 |
ENST00000394894.8
|
KIF20A
|
kinesin family member 20A |
| chr1_+_109113963 | 1.66 |
ENST00000526264.5
|
ELAPOR1
|
endosome-lysosome associated apoptosis and autophagy regulator 1 |
| chr5_+_138179145 | 1.62 |
ENST00000508792.5
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr18_+_12308232 | 1.60 |
ENST00000590103.5
ENST00000591909.5 ENST00000586653.5 ENST00000317702.10 ENST00000592683.5 ENST00000590967.5 ENST00000591208.1 ENST00000591463.1 |
TUBB6
|
tubulin beta 6 class V |
| chr18_+_58671517 | 1.59 |
ENST00000345724.7
|
MALT1
|
MALT1 paracaspase |
| chr10_+_74176537 | 1.56 |
ENST00000672394.1
|
ADK
|
adenosine kinase |
| chr14_-_106374129 | 1.55 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr12_+_93569814 | 1.53 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr10_+_74176741 | 1.49 |
ENST00000673027.1
ENST00000372734.5 ENST00000541550.6 |
ADK
|
adenosine kinase |
| chr2_-_55296361 | 1.48 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr11_-_28108109 | 1.44 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A |
| chr1_+_13585453 | 1.38 |
ENST00000487038.5
ENST00000475043.5 |
PDPN
|
podoplanin |
| chr18_+_58864866 | 1.33 |
ENST00000588456.5
ENST00000591808.6 ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr11_-_82997477 | 1.32 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr12_-_47771029 | 1.30 |
ENST00000549151.5
ENST00000548919.5 |
RAPGEF3
|
Rap guanine nucleotide exchange factor 3 |
| chr12_-_57767057 | 1.30 |
ENST00000228606.9
|
CYP27B1
|
cytochrome P450 family 27 subfamily B member 1 |
| chr10_-_75235917 | 1.28 |
ENST00000469299.1
ENST00000372538.8 |
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr18_-_70205655 | 1.27 |
ENST00000255674.11
ENST00000640769.2 |
RTTN
|
rotatin |
| chr5_+_141402764 | 1.25 |
ENST00000573521.2
ENST00000616887.1 |
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr1_+_86424154 | 1.23 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2 |
| chr19_-_43527189 | 1.21 |
ENST00000292147.7
ENST00000600651.5 |
ETHE1
|
ETHE1 persulfide dioxygenase |
| chr17_+_28335718 | 1.20 |
ENST00000226225.7
|
TNFAIP1
|
TNF alpha induced protein 1 |
| chr19_+_48552159 | 1.19 |
ENST00000201586.7
|
SULT2B1
|
sulfotransferase family 2B member 1 |
| chr19_+_38335775 | 1.18 |
ENST00000410018.5
ENST00000409235.8 ENST00000409410.6 |
CATSPERG
|
cation channel sperm associated auxiliary subunit gamma |
| chr12_+_93570381 | 1.17 |
ENST00000549206.5
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr17_-_59151794 | 1.15 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chrX_+_7219431 | 1.13 |
ENST00000674499.1
ENST00000217961.5 |
STS
|
steroid sulfatase |
| chr20_-_47786553 | 1.08 |
ENST00000467815.5
ENST00000359930.8 ENST00000484875.5 |
SULF2
|
sulfatase 2 |
| chr5_+_157731400 | 1.05 |
ENST00000231198.12
|
THG1L
|
tRNA-histidine guanylyltransferase 1 like |
| chrX_-_132961390 | 1.03 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr15_+_71096941 | 1.03 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr17_+_28335571 | 1.01 |
ENST00000544907.6
|
TNFAIP1
|
TNF alpha induced protein 1 |
| chr11_+_2902258 | 0.96 |
ENST00000649076.2
ENST00000449793.6 |
SLC22A18
|
solute carrier family 22 member 18 |
| chr11_+_126405582 | 0.95 |
ENST00000526727.5
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr14_-_106639589 | 0.95 |
ENST00000390630.3
|
IGHV4-61
|
immunoglobulin heavy variable 4-61 |
| chr10_+_13161543 | 0.94 |
ENST00000378714.8
ENST00000479669.5 ENST00000484800.6 |
MCM10
|
minichromosome maintenance 10 replication initiation factor |
| chr15_-_21742799 | 0.93 |
ENST00000622410.2
|
ENSG00000278263.2
|
novel protein, identical to IGHV4-4 |
| chr17_-_5234801 | 0.93 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr15_+_62066975 | 0.92 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chr3_+_101849505 | 0.90 |
ENST00000326151.9
ENST00000326172.9 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr1_-_38859669 | 0.90 |
ENST00000373001.4
|
RRAGC
|
Ras related GTP binding C |
| chr8_-_63026179 | 0.88 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase |
| chr9_+_127451549 | 0.86 |
ENST00000675883.1
ENST00000676014.1 ENST00000675253.1 |
LRSAM1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr3_-_113746218 | 0.85 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr11_+_31369834 | 0.84 |
ENST00000465995.6
|
DNAJC24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr2_+_191678122 | 0.83 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr7_+_134843884 | 0.82 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr9_+_127451495 | 0.81 |
ENST00000373324.8
ENST00000675572.1 ENST00000676170.1 ENST00000675789.1 ENST00000300417.11 ENST00000675448.1 ENST00000323301.8 ENST00000675141.1 |
LRSAM1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr9_-_137070548 | 0.81 |
ENST00000409687.5
|
SAPCD2
|
suppressor APC domain containing 2 |
| chr11_+_46381753 | 0.80 |
ENST00000407067.1
|
MDK
|
midkine |
| chr18_+_58671439 | 0.79 |
ENST00000649217.2
|
MALT1
|
MALT1 paracaspase |
| chr7_+_127593727 | 0.79 |
ENST00000478821.1
ENST00000265825.6 |
FSCN3
|
fascin actin-bundling protein 3 |
| chr11_+_2902388 | 0.79 |
ENST00000380574.5
|
SLC22A18
|
solute carrier family 22 member 18 |
| chr11_+_46381698 | 0.77 |
ENST00000395565.5
|
MDK
|
midkine |
| chr16_+_56608577 | 0.73 |
ENST00000245185.6
ENST00000561491.1 |
MT2A
|
metallothionein 2A |
| chr5_-_11589019 | 0.73 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr5_+_148063971 | 0.73 |
ENST00000398454.5
ENST00000359874.7 ENST00000508733.5 ENST00000256084.8 |
SPINK5
|
serine peptidase inhibitor Kazal type 5 |
| chr11_+_47248885 | 0.72 |
ENST00000395397.7
ENST00000405576.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr10_-_93482194 | 0.72 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr1_+_111227610 | 0.72 |
ENST00000369744.6
|
CHI3L2
|
chitinase 3 like 2 |
| chr2_+_190927649 | 0.72 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr6_+_63572472 | 0.71 |
ENST00000370651.7
ENST00000626021.3 |
ENSG00000285976.2
PTP4A1
|
novel protein protein tyrosine phosphatase 4A1 |
| chr15_-_22185402 | 0.71 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr10_-_93482326 | 0.70 |
ENST00000359263.9
|
MYOF
|
myoferlin |
| chr4_+_127781815 | 0.70 |
ENST00000508776.5
|
HSPA4L
|
heat shock protein family A (Hsp70) member 4 like |
| chr1_+_117420597 | 0.70 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2 |
| chr11_+_47248924 | 0.69 |
ENST00000481889.6
ENST00000436778.5 ENST00000531660.5 ENST00000407404.5 |
NR1H3
|
nuclear receptor subfamily 1 group H member 3 |
| chr7_+_141790217 | 0.69 |
ENST00000247883.5
|
TAS2R5
|
taste 2 receptor member 5 |
| chr9_-_21995262 | 0.68 |
ENST00000494262.5
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr7_-_86965872 | 0.68 |
ENST00000398276.6
ENST00000416314.5 ENST00000425689.1 |
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr13_+_31945826 | 0.68 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr10_+_10798570 | 0.67 |
ENST00000638035.1
ENST00000636488.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr2_-_171433950 | 0.67 |
ENST00000375258.9
ENST00000442541.1 ENST00000392599.6 |
METTL8
|
methyltransferase like 8 |
| chr6_-_49787265 | 0.66 |
ENST00000304801.6
|
PGK2
|
phosphoglycerate kinase 2 |
| chr11_+_46381645 | 0.66 |
ENST00000617138.4
ENST00000395566.9 ENST00000395569.8 |
MDK
|
midkine |
| chr1_-_212414815 | 0.66 |
ENST00000261455.9
ENST00000535273.2 |
PACC1
|
proton activated chloride channel 1 |
| chr10_+_88594746 | 0.66 |
ENST00000531458.1
|
LIPJ
|
lipase family member J |
| chr22_-_45413589 | 0.66 |
ENST00000357450.9
|
SMC1B
|
structural maintenance of chromosomes 1B |
| chr11_+_47215032 | 0.66 |
ENST00000622090.4
ENST00000378600.7 ENST00000378603.7 |
DDB2
|
damage specific DNA binding protein 2 |
| chr6_+_31971831 | 0.66 |
ENST00000375331.7
ENST00000375333.3 |
STK19
|
serine/threonine kinase 19 |
| chr11_+_5383812 | 0.64 |
ENST00000642046.1
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr14_-_106422175 | 0.64 |
ENST00000390619.2
|
IGHV4-39
|
immunoglobulin heavy variable 4-39 |
| chr11_+_57597563 | 0.63 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chr9_-_21995301 | 0.63 |
ENST00000498628.6
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr4_-_173334249 | 0.62 |
ENST00000506267.1
ENST00000296503.10 |
HMGB2
|
high mobility group box 2 |
| chr4_-_88158605 | 0.61 |
ENST00000237612.8
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr1_+_111227699 | 0.61 |
ENST00000369748.9
|
CHI3L2
|
chitinase 3 like 2 |
| chr22_+_38057200 | 0.60 |
ENST00000404072.7
ENST00000424694.5 |
PICK1
|
protein interacting with PRKCA 1 |
| chr7_+_148339452 | 0.59 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr11_+_695778 | 0.59 |
ENST00000526170.6
ENST00000488769.2 ENST00000397510.9 |
TMEM80
|
transmembrane protein 80 |
| chr15_-_34343112 | 0.59 |
ENST00000557912.1
ENST00000328848.6 |
NOP10
|
NOP10 ribonucleoprotein |
| chr4_-_173334385 | 0.58 |
ENST00000446922.6
|
HMGB2
|
high mobility group box 2 |
| chr11_+_57598184 | 0.58 |
ENST00000677625.1
ENST00000676670.1 |
SERPING1
|
serpin family G member 1 |
| chr10_+_5048748 | 0.58 |
ENST00000602997.5
ENST00000439082.7 |
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr6_+_33620329 | 0.58 |
ENST00000374316.9
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor type 3 |
| chr6_+_29111560 | 0.57 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr4_-_52751399 | 0.57 |
ENST00000440542.1
ENST00000443173.6 |
ERVMER34-1
|
endogenous retrovirus group MER34 member 1, envelope |
| chr22_+_38057371 | 0.57 |
ENST00000437453.5
ENST00000356976.8 |
PICK1
|
protein interacting with PRKCA 1 |
| chr2_-_100417608 | 0.56 |
ENST00000264249.8
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr11_+_695591 | 0.56 |
ENST00000608174.6
ENST00000397512.8 |
TMEM80
|
transmembrane protein 80 |
| chr6_+_99606833 | 0.54 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13 |
| chr7_+_95485898 | 0.53 |
ENST00000428113.5
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr1_+_153357846 | 0.52 |
ENST00000368738.4
|
S100A9
|
S100 calcium binding protein A9 |
| chr9_+_107306459 | 0.51 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr1_-_221742074 | 0.50 |
ENST00000366899.4
|
DUSP10
|
dual specificity phosphatase 10 |
| chr20_+_36461460 | 0.50 |
ENST00000482872.5
ENST00000495241.5 |
DLGAP4
|
DLG associated protein 4 |
| chr11_+_28108248 | 0.50 |
ENST00000406787.7
ENST00000403099.5 ENST00000407364.8 |
METTL15
|
methyltransferase like 15 |
| chr20_-_63572455 | 0.50 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2 |
| chrX_-_106903331 | 0.50 |
ENST00000411805.1
ENST00000276173.5 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr2_-_187513641 | 0.49 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr6_-_33789090 | 0.49 |
ENST00000614475.4
ENST00000293760.10 |
LEMD2
|
LEM domain nuclear envelope protein 2 |
| chr15_-_82699893 | 0.49 |
ENST00000642989.2
|
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr3_-_113746185 | 0.48 |
ENST00000616174.1
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr12_-_118359639 | 0.48 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr1_+_13389632 | 0.48 |
ENST00000376098.4
|
PRAMEF17
|
PRAME family member 17 |
| chr14_-_106335613 | 0.47 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr4_+_86594301 | 0.47 |
ENST00000427191.6
ENST00000411767.7 ENST00000436978.5 ENST00000502971.5 |
PTPN13
|
protein tyrosine phosphatase non-receptor type 13 |
| chr6_+_116461364 | 0.46 |
ENST00000368606.7
ENST00000368605.3 |
CALHM6
|
calcium homeostasis modulator family member 6 |
| chr3_-_113746059 | 0.46 |
ENST00000477813.5
|
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chrX_+_30653478 | 0.46 |
ENST00000378945.7
ENST00000378941.4 |
GK
|
glycerol kinase |
| chr21_+_46286325 | 0.46 |
ENST00000397701.9
ENST00000397694.5 ENST00000329319.7 ENST00000339195.10 ENST00000397692.5 |
YBEY
|
ybeY metalloendoribonuclease |
| chrX_+_106693838 | 0.45 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr1_+_109113910 | 0.45 |
ENST00000531664.5
ENST00000534476.5 |
ELAPOR1
|
endosome-lysosome associated apoptosis and autophagy regulator 1 |
| chrX_+_153179276 | 0.45 |
ENST00000356661.7
|
MAGEA1
|
MAGE family member A1 |
| chr14_+_71933116 | 0.44 |
ENST00000553530.5
ENST00000556437.5 |
RGS6
|
regulator of G protein signaling 6 |
| chr11_+_65787056 | 0.44 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr14_+_74348440 | 0.44 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr5_+_87267792 | 0.44 |
ENST00000274376.11
|
RASA1
|
RAS p21 protein activator 1 |
| chr17_-_7234262 | 0.43 |
ENST00000575756.5
ENST00000575458.5 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_-_18522051 | 0.43 |
ENST00000262809.9
|
ELL
|
elongation factor for RNA polymerase II |
| chrX_-_120559889 | 0.42 |
ENST00000371323.3
|
CUL4B
|
cullin 4B |
| chr17_+_35587239 | 0.42 |
ENST00000621914.4
ENST00000621668.4 ENST00000616681.4 ENST00000612035.4 ENST00000610402.5 ENST00000614600.4 ENST00000590432.5 ENST00000612116.5 |
AP2B1
|
adaptor related protein complex 2 subunit beta 1 |
| chrX_-_48957871 | 0.41 |
ENST00000610466.4
|
OTUD5
|
OTU deubiquitinase 5 |
| chr3_+_150546671 | 0.41 |
ENST00000487799.5
|
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr7_+_74083785 | 0.41 |
ENST00000336180.7
|
LIMK1
|
LIM domain kinase 1 |
| chr7_-_44573895 | 0.40 |
ENST00000258772.10
ENST00000431640.5 |
DDX56
|
DEAD-box helicase 56 |
| chr2_-_89160329 | 0.39 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chrX_+_130401962 | 0.39 |
ENST00000305536.11
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein X-linked 2 |
| chr12_+_92702843 | 0.39 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr12_-_122266410 | 0.39 |
ENST00000451053.3
|
VPS33A
|
VPS33A core subunit of CORVET and HOPS complexes |
| chr1_-_11058839 | 0.39 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr5_+_55160161 | 0.38 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr2_+_210556590 | 0.37 |
ENST00000233072.10
ENST00000619804.1 |
CPS1
|
carbamoyl-phosphate synthase 1 |
| chr2_+_90021567 | 0.37 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr6_+_138404206 | 0.36 |
ENST00000607197.6
ENST00000367697.7 |
HEBP2
|
heme binding protein 2 |
| chr3_+_150546765 | 0.36 |
ENST00000406576.7
ENST00000460851.6 ENST00000482093.5 ENST00000273435.9 |
EIF2A
|
eukaryotic translation initiation factor 2A |
| chr1_-_21783189 | 0.36 |
ENST00000400301.5
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chrX_+_149881141 | 0.36 |
ENST00000535454.5
ENST00000542674.5 ENST00000286482.6 |
MAGEA8
|
MAGE family member A8 |
| chr17_-_7179544 | 0.35 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr6_+_155149109 | 0.35 |
ENST00000456877.6
ENST00000528391.6 |
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr17_-_7179348 | 0.35 |
ENST00000573083.1
ENST00000574388.5 ENST00000269299.8 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr18_+_9475494 | 0.35 |
ENST00000383432.8
|
RALBP1
|
ralA binding protein 1 |
| chr7_-_104207957 | 0.35 |
ENST00000447452.6
ENST00000297431.9 ENST00000626700.1 |
ORC5
|
origin recognition complex subunit 5 |
| chr11_+_131911396 | 0.34 |
ENST00000425719.6
ENST00000374784.5 |
NTM
|
neurotrimin |
| chr15_+_59611776 | 0.34 |
ENST00000396065.3
ENST00000560585.5 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_+_15641280 | 0.34 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr3_-_114179052 | 0.34 |
ENST00000383673.5
ENST00000295881.9 |
DRD3
|
dopamine receptor D3 |
| chr2_+_112645930 | 0.34 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1 |
| chr12_+_92702983 | 0.34 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr12_+_5043873 | 0.33 |
ENST00000252321.5
|
KCNA5
|
potassium voltage-gated channel subfamily A member 5 |
| chr7_-_151736304 | 0.33 |
ENST00000492843.6
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr18_+_9474994 | 0.33 |
ENST00000019317.8
|
RALBP1
|
ralA binding protein 1 |
| chr20_+_36461303 | 0.33 |
ENST00000475894.5
|
DLGAP4
|
DLG associated protein 4 |
| chr15_+_74615808 | 0.32 |
ENST00000395066.9
ENST00000568139.6 ENST00000563297.5 ENST00000568488.6 |
CLK3
|
CDC like kinase 3 |
| chr11_-_66372099 | 0.32 |
ENST00000311161.11
|
SLC29A2
|
solute carrier family 29 member 2 |
| chr20_-_6054026 | 0.32 |
ENST00000378858.5
|
LRRN4
|
leucine rich repeat neuronal 4 |
| chr20_+_11892493 | 0.31 |
ENST00000422390.5
ENST00000618918.4 |
BTBD3
|
BTB domain containing 3 |
| chr7_-_27147774 | 0.31 |
ENST00000222728.3
|
HOXA6
|
homeobox A6 |
| chrX_-_133218345 | 0.31 |
ENST00000310125.5
|
TFDP3
|
transcription factor Dp family member 3 |
| chr1_+_151198536 | 0.30 |
ENST00000349792.9
ENST00000409426.5 ENST00000368888.9 ENST00000441902.6 ENST00000368890.8 ENST00000424999.1 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase type 1 alpha |
| chrX_+_154379203 | 0.30 |
ENST00000369835.3
ENST00000369842.9 |
EMD
|
emerin |
| chr1_-_225999312 | 0.30 |
ENST00000272091.8
|
SDE2
|
SDE2 telomere maintenance homolog |
| chr2_-_61538516 | 0.30 |
ENST00000676771.1
ENST00000677814.1 ENST00000443240.5 ENST00000677556.1 ENST00000676553.1 |
XPO1
|
exportin 1 |
| chrX_+_30653359 | 0.30 |
ENST00000378943.7
ENST00000378946.7 ENST00000427190.6 |
GK
|
glycerol kinase |
| chrX_-_48957548 | 0.30 |
ENST00000376488.8
ENST00000396743.7 ENST00000156084.8 |
OTUD5
|
OTU deubiquitinase 5 |
| chr2_+_108288639 | 0.30 |
ENST00000326853.9
|
SULT1C2
|
sulfotransferase family 1C member 2 |
| chr21_+_29299368 | 0.29 |
ENST00000399921.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr2_-_99254281 | 0.29 |
ENST00000409238.5
ENST00000423800.5 |
LYG2
|
lysozyme g2 |
| chr5_+_174045673 | 0.29 |
ENST00000303177.8
ENST00000519867.5 |
NSG2
|
neuronal vesicle trafficking associated 2 |
| chr1_+_159437845 | 0.28 |
ENST00000642080.1
|
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr1_+_151766655 | 0.28 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chrX_-_53434341 | 0.28 |
ENST00000375298.4
ENST00000375304.9 ENST00000684692.1 ENST00000168216.11 |
HSD17B10
|
hydroxysteroid 17-beta dehydrogenase 10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.8 | 3.1 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.6 | 1.8 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.5 | 10.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.5 | 1.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.5 | 1.4 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.5 | 3.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.4 | 1.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.4 | 1.2 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.4 | 2.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.4 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 2.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 2.2 | GO:0030421 | defecation(GO:0030421) |
| 0.3 | 2.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.3 | 1.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.7 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.3 | 1.0 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 2.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.7 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 0.6 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.2 | 1.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.7 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 0.7 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 0.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 0.5 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 1.4 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.2 | 0.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 0.8 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.4 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 1.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 1.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) |
| 0.1 | 3.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.1 | 0.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.6 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 2.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.7 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.1 | 0.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.4 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.1 | 1.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.3 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.2 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.1 | 0.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.5 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 2.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 1.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 3.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0097045 | activation of blood coagulation via clotting cascade(GO:0002543) phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.0 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.5 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.4 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.4 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:2000035 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 2.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0061364 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.1 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 3.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.9 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.7 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.3 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 1.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.7 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 2.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.4 | 2.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 1.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.6 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.2 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 1.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.9 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.6 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 11.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 3.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 3.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.7 | 9.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 1.8 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.6 | 1.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 10.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 1.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.3 | 1.7 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.3 | 1.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 1.1 | GO:0004773 | steryl-sulfatase activity(GO:0004773) |
| 0.3 | 2.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.2 | 0.9 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 3.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.8 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.3 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.5 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.4 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.1 | 1.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.1 | 0.4 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 1.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 1.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 2.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 6.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 5.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 3.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 2.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |