Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
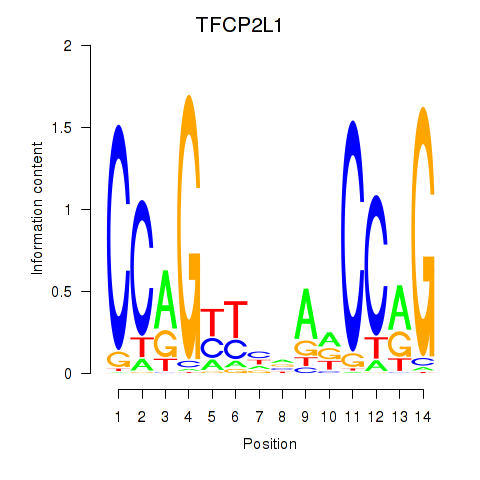
Results for TFCP2L1
Z-value: 0.82
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.8 | TFCP2L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg38_v1_chr2_-_121285194_121285209 | 0.30 | 1.1e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_44651683 | 2.54 |
ENST00000537820.1
ENST00000372874.9 |
ADA
|
adenosine deaminase |
| chr8_-_6877928 | 1.89 |
ENST00000297439.4
|
DEFB1
|
defensin beta 1 |
| chr18_+_23949847 | 1.86 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr2_-_72147819 | 1.69 |
ENST00000001146.7
ENST00000546307.5 ENST00000474509.1 |
CYP26B1
|
cytochrome P450 family 26 subfamily B member 1 |
| chr1_+_116111395 | 1.68 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr2_-_31138429 | 1.64 |
ENST00000349752.10
|
GALNT14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr8_+_53851786 | 1.64 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr12_+_53097656 | 1.63 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr22_+_31093358 | 1.54 |
ENST00000404574.5
|
SMTN
|
smoothelin |
| chr4_+_8199363 | 1.52 |
ENST00000382521.7
ENST00000457650.7 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr10_+_100347225 | 1.51 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase |
| chr4_+_8199239 | 1.51 |
ENST00000245105.8
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr2_+_11746576 | 1.42 |
ENST00000256720.6
ENST00000674199.1 ENST00000441684.5 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr12_-_119803383 | 1.42 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr12_+_40692413 | 1.36 |
ENST00000551295.7
ENST00000547702.5 ENST00000551424.5 |
CNTN1
|
contactin 1 |
| chr11_-_62556230 | 1.29 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr9_+_130053706 | 1.23 |
ENST00000372410.7
|
GPR107
|
G protein-coupled receptor 107 |
| chr21_-_15064934 | 1.18 |
ENST00000400199.5
ENST00000400202.5 ENST00000318948.7 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr4_-_121823843 | 1.10 |
ENST00000274026.10
|
CCNA2
|
cyclin A2 |
| chr7_-_128405930 | 1.07 |
ENST00000470772.5
ENST00000480861.5 ENST00000496200.5 |
IMPDH1
|
inosine monophosphate dehydrogenase 1 |
| chr12_+_119178920 | 1.05 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr12_+_119178953 | 1.03 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr20_+_45881218 | 1.03 |
ENST00000372523.1
|
ZSWIM1
|
zinc finger SWIM-type containing 1 |
| chr19_+_47713412 | 1.02 |
ENST00000538399.1
ENST00000263277.8 |
EHD2
|
EH domain containing 2 |
| chr1_-_155990062 | 1.02 |
ENST00000462460.6
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr19_-_42412347 | 1.01 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase E, hormone sensitive type |
| chr20_-_54173976 | 1.00 |
ENST00000216862.8
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1 |
| chr9_-_124507382 | 0.93 |
ENST00000373588.9
ENST00000620110.4 |
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr22_-_37427433 | 0.92 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr11_+_111245725 | 0.91 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chrX_-_107717054 | 0.88 |
ENST00000503515.1
ENST00000372397.6 |
TSC22D3
|
TSC22 domain family member 3 |
| chr8_+_49072335 | 0.87 |
ENST00000399653.8
ENST00000522267.6 ENST00000303202.8 |
PPDPFL
|
pancreatic progenitor cell differentiation and proliferation factor like |
| chr14_-_106025628 | 0.86 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr12_-_121274510 | 0.86 |
ENST00000392474.6
|
CAMKK2
|
calcium/calmodulin dependent protein kinase kinase 2 |
| chr2_+_11724333 | 0.82 |
ENST00000425416.6
ENST00000396097.5 |
LPIN1
|
lipin 1 |
| chr1_-_26354080 | 0.81 |
ENST00000308182.10
|
CRYBG2
|
crystallin beta-gamma domain containing 2 |
| chr11_+_78188871 | 0.79 |
ENST00000528910.5
ENST00000529308.6 |
USP35
|
ubiquitin specific peptidase 35 |
| chr16_+_46884323 | 0.79 |
ENST00000340124.9
|
GPT2
|
glutamic--pyruvic transaminase 2 |
| chr19_+_14583076 | 0.79 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr19_-_46413548 | 0.76 |
ENST00000307522.5
|
CCDC8
|
coiled-coil domain containing 8 |
| chr11_+_62881686 | 0.74 |
ENST00000536981.6
ENST00000539891.6 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr1_-_161550591 | 0.73 |
ENST00000367967.7
ENST00000436743.6 ENST00000442336.1 |
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr14_-_68937942 | 0.73 |
ENST00000684182.1
|
ACTN1
|
actinin alpha 1 |
| chr11_+_95089804 | 0.72 |
ENST00000278505.5
|
ENDOD1
|
endonuclease domain containing 1 |
| chr17_-_76027212 | 0.71 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr7_+_151956379 | 0.69 |
ENST00000431418.6
|
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chrX_-_64205680 | 0.67 |
ENST00000374869.8
|
AMER1
|
APC membrane recruitment protein 1 |
| chr7_+_151956440 | 0.67 |
ENST00000392800.7
ENST00000616416.4 |
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chr14_+_64504574 | 0.66 |
ENST00000358738.3
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_-_161549793 | 0.66 |
ENST00000443193.6
|
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr1_-_161549892 | 0.66 |
ENST00000426740.7
|
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr19_-_1822038 | 0.66 |
ENST00000643515.1
|
REXO1
|
RNA exonuclease 1 homolog |
| chr2_+_90234809 | 0.65 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr17_-_76027296 | 0.65 |
ENST00000301607.8
|
EVPL
|
envoplakin |
| chr19_+_41219177 | 0.65 |
ENST00000301178.9
|
AXL
|
AXL receptor tyrosine kinase |
| chr15_+_74615808 | 0.63 |
ENST00000395066.9
ENST00000568139.6 ENST00000563297.5 ENST00000568488.6 |
CLK3
|
CDC like kinase 3 |
| chr9_+_130053897 | 0.61 |
ENST00000347136.11
ENST00000610997.1 |
GPR107
|
G protein-coupled receptor 107 |
| chr17_+_40015428 | 0.60 |
ENST00000394149.8
ENST00000225474.6 ENST00000331769.6 ENST00000394148.7 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 |
| chr14_-_68979076 | 0.57 |
ENST00000538545.6
ENST00000684639.1 |
ACTN1
|
actinin alpha 1 |
| chr1_+_11934651 | 0.57 |
ENST00000449038.5
ENST00000196061.5 ENST00000429000.6 |
PLOD1
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 |
| chrX_-_50200988 | 0.57 |
ENST00000358526.7
|
AKAP4
|
A-kinase anchoring protein 4 |
| chr14_-_68979251 | 0.57 |
ENST00000438964.6
ENST00000679147.1 |
ACTN1
|
actinin alpha 1 |
| chr4_-_993430 | 0.56 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr2_+_71453538 | 0.56 |
ENST00000258104.8
|
DYSF
|
dysferlin |
| chr1_-_201115372 | 0.55 |
ENST00000458416.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr14_-_68979274 | 0.55 |
ENST00000394419.9
|
ACTN1
|
actinin alpha 1 |
| chr10_-_119536533 | 0.55 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr14_-_68979314 | 0.52 |
ENST00000684713.1
ENST00000683198.1 ENST00000684598.1 ENST00000682331.1 ENST00000682291.1 ENST00000683342.1 |
ACTN1
|
actinin alpha 1 |
| chr17_+_18377769 | 0.52 |
ENST00000399134.5
|
EVPLL
|
envoplakin like |
| chr2_+_233251694 | 0.52 |
ENST00000417017.5
ENST00000392020.8 ENST00000392018.1 |
ATG16L1
|
autophagy related 16 like 1 |
| chr15_-_82699893 | 0.51 |
ENST00000642989.2
|
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr14_-_68979436 | 0.51 |
ENST00000193403.10
|
ACTN1
|
actinin alpha 1 |
| chr17_-_1492660 | 0.50 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr7_+_77538059 | 0.50 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr7_-_100158679 | 0.50 |
ENST00000456769.5
ENST00000316937.8 |
TRAPPC14
|
trafficking protein particle complex 14 |
| chr12_-_53232182 | 0.50 |
ENST00000425354.7
ENST00000546717.1 ENST00000394426.5 |
RARG
|
retinoic acid receptor gamma |
| chr2_+_233251571 | 0.50 |
ENST00000347464.9
ENST00000444735.5 ENST00000373525.9 ENST00000392017.9 ENST00000419681.5 |
ATG16L1
|
autophagy related 16 like 1 |
| chr18_+_50560070 | 0.49 |
ENST00000400384.7
ENST00000540640.3 ENST00000592595.5 |
MAPK4
|
mitogen-activated protein kinase 4 |
| chr3_+_26622800 | 0.48 |
ENST00000396641.6
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr1_-_155978524 | 0.47 |
ENST00000361247.9
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr1_-_161631032 | 0.47 |
ENST00000534776.1
ENST00000613418.4 ENST00000614870.4 |
FCGR3B
|
Fc fragment of IgG receptor IIIb |
| chr5_+_140360187 | 0.47 |
ENST00000506757.7
ENST00000506545.5 ENST00000432095.6 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4 member 9 |
| chr11_-_86068743 | 0.46 |
ENST00000356360.9
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr22_+_31160171 | 0.46 |
ENST00000326132.11
ENST00000426256.6 ENST00000266252.8 |
RNF185
|
ring finger protein 185 |
| chr1_-_161631152 | 0.46 |
ENST00000421702.3
ENST00000650385.1 |
FCGR3B
|
Fc fragment of IgG receptor IIIb |
| chr5_-_152405277 | 0.46 |
ENST00000255262.4
|
NMUR2
|
neuromedin U receptor 2 |
| chr14_-_106154113 | 0.46 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr11_-_86068929 | 0.45 |
ENST00000630913.2
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_+_62613236 | 0.44 |
ENST00000278833.4
|
ROM1
|
retinal outer segment membrane protein 1 |
| chr22_-_38088915 | 0.44 |
ENST00000428572.1
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chrX_-_132218124 | 0.43 |
ENST00000342983.6
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr12_+_55931148 | 0.43 |
ENST00000549629.5
ENST00000555218.5 ENST00000331886.10 |
DGKA
|
diacylglycerol kinase alpha |
| chr19_+_38930916 | 0.42 |
ENST00000308018.9
ENST00000407800.2 ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr10_+_97589715 | 0.42 |
ENST00000370640.5
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr9_-_34637719 | 0.41 |
ENST00000378892.5
ENST00000680277.1 ENST00000277010.9 ENST00000679597.1 ENST00000680244.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr19_+_40717091 | 0.41 |
ENST00000263370.3
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr5_-_36241798 | 0.41 |
ENST00000514504.5
|
NADK2
|
NAD kinase 2, mitochondrial |
| chr5_-_177496845 | 0.41 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr3_-_50340804 | 0.40 |
ENST00000359365.9
ENST00000357043.6 |
RASSF1
|
Ras association domain family member 1 |
| chr10_+_122163590 | 0.40 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr3_-_50340933 | 0.39 |
ENST00000616212.4
|
RASSF1
|
Ras association domain family member 1 |
| chr3_-_48635426 | 0.39 |
ENST00000455886.6
ENST00000431739.5 ENST00000426599.1 ENST00000383733.7 ENST00000395550.7 ENST00000420764.6 ENST00000337000.12 |
SLC26A6
|
solute carrier family 26 member 6 |
| chr10_+_104641282 | 0.39 |
ENST00000369701.8
|
SORCS3
|
sortilin related VPS10 domain containing receptor 3 |
| chr22_-_38755458 | 0.39 |
ENST00000405510.5
ENST00000433561.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr4_+_40193642 | 0.39 |
ENST00000617441.4
ENST00000503941.5 |
RHOH
|
ras homolog family member H |
| chr2_+_71453722 | 0.39 |
ENST00000409582.7
ENST00000409762.5 ENST00000413539.6 ENST00000429174.6 |
DYSF
|
dysferlin |
| chr5_-_177496802 | 0.39 |
ENST00000506161.5
|
PDLIM7
|
PDZ and LIM domain 7 |
| chr6_-_3751703 | 0.38 |
ENST00000380283.5
|
PXDC1
|
PX domain containing 1 |
| chr20_-_31870510 | 0.38 |
ENST00000339738.10
|
DUSP15
|
dual specificity phosphatase 15 |
| chr2_+_9206762 | 0.38 |
ENST00000315273.4
ENST00000281419.8 |
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr8_-_53842899 | 0.38 |
ENST00000524234.1
ENST00000521275.5 ENST00000396774.6 |
ATP6V1H
|
ATPase H+ transporting V1 subunit H |
| chr12_-_120369156 | 0.37 |
ENST00000257552.7
|
MSI1
|
musashi RNA binding protein 1 |
| chr9_-_34637800 | 0.36 |
ENST00000680730.1
ENST00000477726.1 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr19_-_18543483 | 0.36 |
ENST00000597547.1
ENST00000222308.8 |
FKBP8
|
FKBP prolyl isomerase 8 |
| chr1_+_151540299 | 0.36 |
ENST00000392712.7
ENST00000368848.6 ENST00000368849.8 ENST00000353024.4 |
TUFT1
|
tuftelin 1 |
| chr2_-_168247569 | 0.36 |
ENST00000355999.5
|
STK39
|
serine/threonine kinase 39 |
| chr17_+_7355123 | 0.36 |
ENST00000389982.8
ENST00000576060.6 ENST00000330767.4 |
TMEM95
|
transmembrane protein 95 |
| chr19_+_33621944 | 0.36 |
ENST00000650847.1
ENST00000591231.5 ENST00000434302.5 ENST00000438847.7 |
CHST8
|
carbohydrate sulfotransferase 8 |
| chr12_-_48106042 | 0.35 |
ENST00000551798.1
ENST00000549518.6 |
SENP1
|
SUMO specific peptidase 1 |
| chr10_+_122163426 | 0.35 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr19_+_4909430 | 0.35 |
ENST00000620565.4
ENST00000613817.4 ENST00000624301.3 ENST00000650932.1 |
UHRF1
|
ubiquitin like with PHD and ring finger domains 1 |
| chr3_-_151316795 | 0.35 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr19_-_18542971 | 0.35 |
ENST00000596558.6
|
FKBP8
|
FKBP prolyl isomerase 8 |
| chr7_+_74083785 | 0.35 |
ENST00000336180.7
|
LIMK1
|
LIM domain kinase 1 |
| chr1_-_156248038 | 0.35 |
ENST00000470198.5
ENST00000292291.10 ENST00000356983.7 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr19_+_45251363 | 0.35 |
ENST00000620044.4
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr4_-_163332589 | 0.35 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr8_-_116766255 | 0.34 |
ENST00000276682.8
|
EIF3H
|
eukaryotic translation initiation factor 3 subunit H |
| chr17_-_45410414 | 0.34 |
ENST00000532038.5
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr19_-_9818808 | 0.33 |
ENST00000586651.5
ENST00000586073.1 |
FBXL12
|
F-box and leucine rich repeat protein 12 |
| chr15_+_75724034 | 0.33 |
ENST00000332145.3
|
ODF3L1
|
outer dense fiber of sperm tails 3 like 1 |
| chr3_-_48188356 | 0.33 |
ENST00000351231.7
ENST00000437972.1 ENST00000302506.8 |
CDC25A
|
cell division cycle 25A |
| chr19_+_18612848 | 0.32 |
ENST00000262817.8
|
TMEM59L
|
transmembrane protein 59 like |
| chr10_+_122163672 | 0.32 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr2_+_231781669 | 0.32 |
ENST00000410024.5
ENST00000611614.4 ENST00000409295.5 ENST00000409091.5 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr5_+_141475928 | 0.32 |
ENST00000611950.1
ENST00000308177.5 ENST00000617641.4 ENST00000621008.1 ENST00000617222.4 |
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr2_+_181891974 | 0.32 |
ENST00000409001.5
|
ITPRID2
|
ITPR interacting domain containing 2 |
| chr14_+_92652468 | 0.32 |
ENST00000556418.1
|
RIN3
|
Ras and Rab interactor 3 |
| chr19_+_48465837 | 0.32 |
ENST00000595676.1
|
ENSG00000268465.1
|
novel protein |
| chr19_+_45251249 | 0.32 |
ENST00000262891.9
ENST00000300843.8 |
MARK4
|
microtubule affinity regulating kinase 4 |
| chr19_+_41354145 | 0.32 |
ENST00000604123.5
|
TMEM91
|
transmembrane protein 91 |
| chr12_+_48106094 | 0.31 |
ENST00000546755.5
ENST00000549366.5 ENST00000642730.1 ENST00000552792.5 |
PFKM
|
phosphofructokinase, muscle |
| chr1_-_211133945 | 0.31 |
ENST00000640044.1
ENST00000640566.1 |
KCNH1
|
potassium voltage-gated channel subfamily H member 1 |
| chr10_+_100745711 | 0.31 |
ENST00000370296.6
ENST00000428433.5 |
PAX2
|
paired box 2 |
| chr8_-_97277890 | 0.31 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5 |
| chr1_-_156248013 | 0.31 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr19_-_20661507 | 0.30 |
ENST00000612591.4
ENST00000595405.1 |
ZNF626
|
zinc finger protein 626 |
| chr1_+_26863140 | 0.30 |
ENST00000339276.6
|
SFN
|
stratifin |
| chr13_+_20703677 | 0.30 |
ENST00000682841.1
|
IL17D
|
interleukin 17D |
| chr17_+_7420315 | 0.30 |
ENST00000323675.4
|
SPEM1
|
spermatid maturation 1 |
| chr6_-_31795627 | 0.30 |
ENST00000375663.8
|
VARS1
|
valyl-tRNA synthetase 1 |
| chr19_-_9819032 | 0.29 |
ENST00000590277.1
ENST00000588922.5 ENST00000589626.5 ENST00000247977.9 ENST00000592067.1 ENST00000586469.1 |
FBXL12
|
F-box and leucine rich repeat protein 12 |
| chr5_+_96663010 | 0.29 |
ENST00000506811.5
ENST00000514055.5 ENST00000508608.6 |
CAST
|
calpastatin |
| chr17_-_16569184 | 0.29 |
ENST00000448349.2
ENST00000395825.4 |
ZNF287
|
zinc finger protein 287 |
| chr17_+_42844573 | 0.29 |
ENST00000253799.8
ENST00000452774.2 |
AOC2
|
amine oxidase copper containing 2 |
| chr2_+_26764232 | 0.29 |
ENST00000344420.10
|
SLC35F6
|
solute carrier family 35 member F6 |
| chr2_+_181891697 | 0.28 |
ENST00000431877.7
|
ITPRID2
|
ITPR interacting domain containing 2 |
| chr2_+_181891904 | 0.28 |
ENST00000320370.11
|
ITPRID2
|
ITPR interacting domain containing 2 |
| chr8_-_27992624 | 0.28 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5 |
| chr2_+_102619531 | 0.28 |
ENST00000233969.3
|
SLC9A2
|
solute carrier family 9 member A2 |
| chr15_-_89690775 | 0.28 |
ENST00000559170.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr10_+_63133247 | 0.28 |
ENST00000435510.6
|
NRBF2
|
nuclear receptor binding factor 2 |
| chr17_-_41027198 | 0.28 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr19_-_6375849 | 0.27 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr15_-_89690634 | 0.27 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr1_-_207032749 | 0.27 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr17_-_16569169 | 0.27 |
ENST00000395824.5
|
ZNF287
|
zinc finger protein 287 |
| chr4_+_146638890 | 0.26 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2 |
| chr10_+_63133279 | 0.26 |
ENST00000277746.11
|
NRBF2
|
nuclear receptor binding factor 2 |
| chr4_-_10116779 | 0.26 |
ENST00000499869.7
|
WDR1
|
WD repeat domain 1 |
| chr15_+_60004305 | 0.26 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr5_-_141619049 | 0.26 |
ENST00000647433.1
ENST00000253811.10 ENST00000389057.9 ENST00000398557.8 |
DIAPH1
|
diaphanous related formin 1 |
| chr12_+_93574965 | 0.26 |
ENST00000551883.1
ENST00000549510.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr6_+_31587185 | 0.25 |
ENST00000376092.7
ENST00000376086.7 ENST00000303757.12 ENST00000376093.6 |
LST1
|
leukocyte specific transcript 1 |
| chr10_+_100745570 | 0.25 |
ENST00000427256.6
ENST00000355243.8 |
PAX2
|
paired box 2 |
| chr5_+_150671588 | 0.25 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr22_+_20131781 | 0.25 |
ENST00000334554.12
ENST00000320602.11 ENST00000405930.3 |
ZDHHC8
|
zinc finger DHHC-type palmitoyltransferase 8 |
| chr11_+_18322253 | 0.25 |
ENST00000453096.6
|
GTF2H1
|
general transcription factor IIH subunit 1 |
| chr3_+_184362599 | 0.25 |
ENST00000455712.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr14_-_106803221 | 0.25 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr19_+_10602436 | 0.24 |
ENST00000590382.5
ENST00000407327.8 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr1_-_161132577 | 0.24 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr1_+_20290869 | 0.24 |
ENST00000289815.13
ENST00000375079.6 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr12_+_57772648 | 0.24 |
ENST00000333012.5
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr20_+_45406035 | 0.23 |
ENST00000372723.7
ENST00000372722.7 |
DBNDD2
|
dysbindin domain containing 2 |
| chr2_-_46941710 | 0.23 |
ENST00000409207.5
|
MCFD2
|
multiple coagulation factor deficiency 2, ER cargo receptor complex subunit |
| chr20_+_63235899 | 0.23 |
ENST00000217169.8
|
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr5_-_141618957 | 0.23 |
ENST00000389054.8
|
DIAPH1
|
diaphanous related formin 1 |
| chr5_-_141618914 | 0.23 |
ENST00000518047.5
|
DIAPH1
|
diaphanous related formin 1 |
| chr2_-_20012661 | 0.23 |
ENST00000421259.2
ENST00000407540.8 |
MATN3
|
matrilin 3 |
| chr12_+_57772587 | 0.22 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr11_-_86069043 | 0.22 |
ENST00000532317.5
ENST00000528256.1 ENST00000393346.8 ENST00000526033.5 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr4_+_56435730 | 0.22 |
ENST00000514888.5
ENST00000264221.6 ENST00000505164.5 |
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr10_+_100746197 | 0.22 |
ENST00000361791.7
|
PAX2
|
paired box 2 |
| chr2_+_71068636 | 0.22 |
ENST00000244204.11
ENST00000533981.5 |
NAGK
|
N-acetylglucosamine kinase |
| chr6_-_31582415 | 0.22 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr3_-_71753582 | 0.22 |
ENST00000295612.7
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr2_-_27071628 | 0.22 |
ENST00000447619.5
ENST00000429985.1 ENST00000456793.2 |
OST4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr3_+_52287811 | 0.22 |
ENST00000305690.12
ENST00000473032.5 ENST00000436784.7 ENST00000471180.5 |
GLYCTK
|
glycerate kinase |
| chr4_+_56436131 | 0.21 |
ENST00000399688.7
|
PAICS
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0042323 | germinal center B cell differentiation(GO:0002314) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) xanthine metabolic process(GO:0046110) negative regulation of mucus secretion(GO:0070256) |
| 0.6 | 1.7 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.5 | 1.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.4 | 1.1 | GO:0071373 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.3 | 2.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 1.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 1.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.2 | 0.9 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 0.8 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.2 | 1.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 1.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 0.8 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.2 | 0.5 | GO:0043006 | reduction of food intake in response to dietary excess(GO:0002023) activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.9 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.6 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 3.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.5 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.7 | GO:2000669 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 0.3 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.1 | 0.4 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 0.6 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.7 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.4 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.2 | GO:0060913 | inner cell mass cellular morphogenesis(GO:0001828) cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.6 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.4 | GO:2000371 | regulation of methylation-dependent chromatin silencing(GO:0090308) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) |
| 0.0 | 0.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.5 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.2 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 1.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 2.6 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 1.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.6 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 1.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) positive regulation of nuclease activity(GO:0032075) positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:1990742 | microvesicle(GO:1990742) |
| 0.3 | 1.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.6 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 3.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.8 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 1.1 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 2.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 1.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.6 | 1.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 1.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.3 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 0.8 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.2 | 0.7 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.2 | 1.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 3.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 3.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0004638 | phosphoribosylaminoimidazole carboxylase activity(GO:0004638) phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.6 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 1.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.8 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 3.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 2.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.4 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.4 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 1.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.4 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 0.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 1.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 1.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.1 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 2.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 3.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |