Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for THRA_RXRB
Z-value: 0.81
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
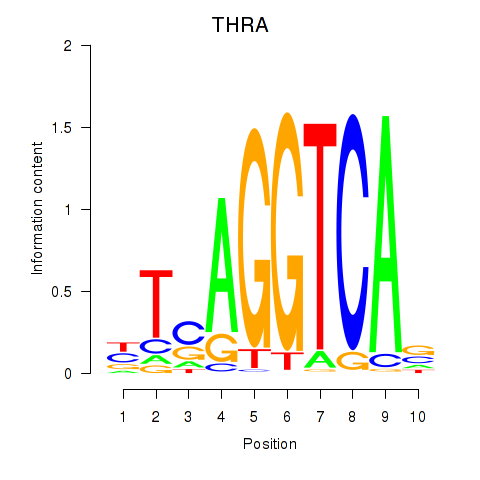
THRA
|
ENSG00000126351.13 | THRA |
|
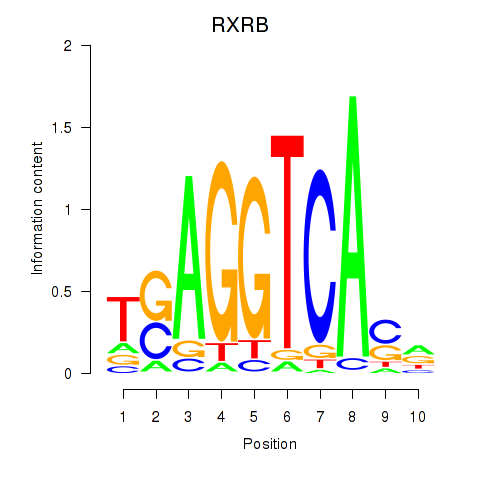
RXRB
|
ENSG00000204231.11 | RXRB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRA | hg38_v1_chr17_+_40062810_40062901 | -0.61 | 3.6e-04 | Click! |
| RXRB | hg38_v1_chr6_-_33200614_33200680 | -0.48 | 7.5e-03 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_152908538 | 3.44 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr5_+_163460623 | 2.54 |
ENST00000393915.9
ENST00000432118.6 |
HMMR
|
hyaluronan mediated motility receptor |
| chr10_+_11742361 | 2.30 |
ENST00000379215.9
ENST00000420401.5 |
ECHDC3
|
enoyl-CoA hydratase domain containing 3 |
| chr19_-_51065067 | 2.21 |
ENST00000595547.5
ENST00000335422.3 ENST00000595793.6 ENST00000596955.1 |
KLK13
|
kallikrein related peptidase 13 |
| chr1_+_150549734 | 2.16 |
ENST00000674043.1
ENST00000674058.1 |
ADAMTSL4
|
ADAMTS like 4 |
| chr19_+_53867874 | 2.09 |
ENST00000448420.5
ENST00000439000.5 ENST00000391771.1 ENST00000391770.9 |
MYADM
|
myeloid associated differentiation marker |
| chr1_-_150720842 | 1.98 |
ENST00000442853.5
ENST00000368995.8 ENST00000322343.11 ENST00000361824.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr22_+_22922594 | 1.95 |
ENST00000390331.3
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr9_+_122371036 | 1.93 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr15_-_34337719 | 1.93 |
ENST00000559484.1
ENST00000558589.5 ENST00000458406.6 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr15_-_34338033 | 1.78 |
ENST00000558667.5
ENST00000561120.5 ENST00000559236.5 ENST00000397702.6 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr3_-_50303565 | 1.64 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr15_-_34337772 | 1.63 |
ENST00000354181.8
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr1_+_44746401 | 1.58 |
ENST00000372217.5
|
KIF2C
|
kinesin family member 2C |
| chr12_-_95116967 | 1.52 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr22_-_37188281 | 1.46 |
ENST00000397110.6
|
C1QTNF6
|
C1q and TNF related 6 |
| chr19_+_6739650 | 1.44 |
ENST00000313285.12
ENST00000313244.14 ENST00000596758.5 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr1_+_17205119 | 1.42 |
ENST00000375471.5
|
PADI1
|
peptidyl arginine deiminase 1 |
| chr5_+_148312416 | 1.40 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr19_+_16076485 | 1.36 |
ENST00000643579.2
ENST00000646575.1 |
TPM4
|
tropomyosin 4 |
| chr19_+_44777860 | 1.35 |
ENST00000341505.4
ENST00000647358.2 |
CBLC
|
Cbl proto-oncogene C |
| chr19_-_51723968 | 1.33 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr3_+_12287859 | 1.32 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr9_+_113536497 | 1.30 |
ENST00000462143.5
|
RGS3
|
regulator of G protein signaling 3 |
| chr2_-_191151568 | 1.27 |
ENST00000358470.8
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr3_+_10026409 | 1.27 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chr3_+_12287962 | 1.25 |
ENST00000643197.2
ENST00000644622.2 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_12288838 | 1.25 |
ENST00000455517.6
ENST00000681982.1 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_12287899 | 1.23 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr22_-_28741783 | 1.23 |
ENST00000439200.5
ENST00000405598.5 ENST00000398017.3 ENST00000649563.1 ENST00000650281.1 ENST00000425190.7 ENST00000404276.6 ENST00000650233.1 ENST00000348295.7 ENST00000382580.6 |
CHEK2
|
checkpoint kinase 2 |
| chr20_+_44715360 | 1.21 |
ENST00000190983.5
|
CCN5
|
cellular communication network factor 5 |
| chr3_+_12289061 | 1.17 |
ENST00000652522.1
ENST00000652431.1 ENST00000652098.1 ENST00000651735.1 ENST00000397026.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr12_+_119178920 | 1.17 |
ENST00000281938.7
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr4_-_110198579 | 1.13 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_110198650 | 1.12 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_+_119178953 | 1.12 |
ENST00000674542.1
|
HSPB8
|
heat shock protein family B (small) member 8 |
| chr10_-_96357155 | 1.08 |
ENST00000536387.5
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr1_-_175192911 | 1.07 |
ENST00000444639.5
|
KIAA0040
|
KIAA0040 |
| chr3_-_197298558 | 1.02 |
ENST00000656944.1
ENST00000346964.6 ENST00000448528.6 ENST00000655488.1 ENST00000357674.9 ENST00000667157.1 ENST00000661336.1 ENST00000654737.1 ENST00000659716.1 ENST00000657381.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr17_-_29176752 | 1.00 |
ENST00000533112.5
|
MYO18A
|
myosin XVIIIA |
| chr8_-_143568854 | 1.00 |
ENST00000524906.5
ENST00000532862.1 ENST00000534459.5 |
MROH6
|
maestro heat like repeat family member 6 |
| chrX_+_136148440 | 0.99 |
ENST00000627383.2
ENST00000630084.2 |
FHL1
|
four and a half LIM domains 1 |
| chr20_+_3796288 | 0.98 |
ENST00000439880.6
ENST00000245960.10 |
CDC25B
|
cell division cycle 25B |
| chr11_-_58575846 | 0.97 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr9_+_36572854 | 0.96 |
ENST00000543751.5
ENST00000536860.5 ENST00000541717.4 ENST00000536329.5 ENST00000536987.5 ENST00000298048.7 ENST00000495529.5 ENST00000545008.5 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr1_-_175192769 | 0.91 |
ENST00000423313.6
|
KIAA0040
|
KIAA0040 |
| chr22_-_37188233 | 0.90 |
ENST00000434784.1
ENST00000337843.7 |
C1QTNF6
|
C1q and TNF related 6 |
| chr5_-_132227808 | 0.79 |
ENST00000401867.5
ENST00000379086.5 ENST00000379100.7 ENST00000418055.5 ENST00000453286.5 ENST00000360568.8 ENST00000379104.7 ENST00000166534.8 |
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr12_+_102120240 | 0.78 |
ENST00000537257.5
ENST00000392911.6 |
PARPBP
|
PARP1 binding protein |
| chr22_-_21227637 | 0.76 |
ENST00000401924.5
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr11_-_123885627 | 0.76 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr21_-_31558977 | 0.75 |
ENST00000286827.7
ENST00000541036.5 |
TIAM1
|
TIAM Rac1 associated GEF 1 |
| chr12_+_102120172 | 0.75 |
ENST00000327680.7
ENST00000541394.5 ENST00000543784.5 |
PARPBP
|
PARP1 binding protein |
| chr17_-_5234801 | 0.73 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr22_+_22900976 | 0.73 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 |
| chr1_+_155613221 | 0.73 |
ENST00000462250.2
|
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr16_+_67666770 | 0.71 |
ENST00000403458.9
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr2_+_209653171 | 0.70 |
ENST00000447185.5
|
MAP2
|
microtubule associated protein 2 |
| chr1_-_159925496 | 0.69 |
ENST00000368097.9
|
TAGLN2
|
transgelin 2 |
| chr19_-_40690629 | 0.69 |
ENST00000252891.8
|
NUMBL
|
NUMB like endocytic adaptor protein |
| chr5_-_132227472 | 0.68 |
ENST00000428369.6
|
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr6_+_35805347 | 0.64 |
ENST00000360215.3
|
LHFPL5
|
LHFPL tetraspan subfamily member 5 |
| chr15_-_34337462 | 0.64 |
ENST00000676379.1
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr22_-_37484505 | 0.63 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_150982268 | 0.63 |
ENST00000368947.9
|
ANXA9
|
annexin A9 |
| chr2_+_218607861 | 0.62 |
ENST00000450993.7
|
PLCD4
|
phospholipase C delta 4 |
| chr16_-_58295019 | 0.61 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr1_-_6393750 | 0.60 |
ENST00000545482.5
ENST00000361521.9 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr16_-_58294976 | 0.59 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr1_+_70411180 | 0.58 |
ENST00000411986.6
|
CTH
|
cystathionine gamma-lyase |
| chrX_+_102125703 | 0.58 |
ENST00000329035.2
|
TCEAL2
|
transcription elongation factor A like 2 |
| chrX_+_102125668 | 0.58 |
ENST00000372780.6
|
TCEAL2
|
transcription elongation factor A like 2 |
| chr12_+_78863962 | 0.57 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1 |
| chr14_-_105601728 | 0.55 |
ENST00000641420.1
ENST00000390541.2 |
IGHE
|
immunoglobulin heavy constant epsilon |
| chr7_+_2244476 | 0.53 |
ENST00000397049.2
|
NUDT1
|
nudix hydrolase 1 |
| chr1_+_70411241 | 0.53 |
ENST00000370938.8
ENST00000346806.2 |
CTH
|
cystathionine gamma-lyase |
| chr1_+_154327737 | 0.53 |
ENST00000672630.1
|
ATP8B2
|
ATPase phospholipid transporting 8B2 |
| chr22_+_18529810 | 0.52 |
ENST00000613597.1
|
TMEM191B
|
transmembrane protein 191B |
| chr2_+_130182224 | 0.52 |
ENST00000651060.1
ENST00000409255.1 ENST00000281871.11 ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr4_-_1856090 | 0.51 |
ENST00000302787.3
|
LETM1
|
leucine zipper and EF-hand containing transmembrane protein 1 |
| chr1_-_6393339 | 0.51 |
ENST00000608083.5
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr10_-_114404480 | 0.51 |
ENST00000419268.1
ENST00000304129.9 |
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr17_-_41060109 | 0.50 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr15_-_45129857 | 0.50 |
ENST00000267803.8
ENST00000613425.4 ENST00000559014.5 ENST00000558851.1 ENST00000559988.5 ENST00000558996.5 ENST00000558422.5 ENST00000559226.5 ENST00000560572.6 ENST00000558326.5 ENST00000558377.5 ENST00000559644.5 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr1_-_243843164 | 0.49 |
ENST00000491219.6
ENST00000680056.1 ENST00000492957.2 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr9_-_111036207 | 0.49 |
ENST00000541779.5
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr10_-_114404756 | 0.49 |
ENST00000369271.7
|
AFAP1L2
|
actin filament associated protein 1 like 2 |
| chr1_+_24745396 | 0.49 |
ENST00000374379.9
|
CLIC4
|
chloride intracellular channel 4 |
| chr1_-_205935822 | 0.48 |
ENST00000340781.8
|
SLC26A9
|
solute carrier family 26 member 9 |
| chr8_+_27774566 | 0.48 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr10_-_75099489 | 0.47 |
ENST00000472493.6
ENST00000478873.7 ENST00000605915.5 |
DUSP13
|
dual specificity phosphatase 13 |
| chrX_-_132961390 | 0.46 |
ENST00000370836.6
ENST00000521489.5 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr20_+_63861498 | 0.46 |
ENST00000369916.5
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr17_+_39738317 | 0.45 |
ENST00000394211.7
|
GRB7
|
growth factor receptor bound protein 7 |
| chr22_-_23974441 | 0.45 |
ENST00000398344.9
ENST00000403754.7 ENST00000430101.2 |
DDT
|
D-dopachrome tautomerase |
| chr5_+_136058849 | 0.44 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr13_+_75549477 | 0.43 |
ENST00000618773.4
|
UCHL3
|
ubiquitin C-terminal hydrolase L3 |
| chr10_+_26697653 | 0.43 |
ENST00000376215.10
|
PDSS1
|
decaprenyl diphosphate synthase subunit 1 |
| chr14_-_105708627 | 0.43 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr18_+_63476927 | 0.43 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr17_-_1492660 | 0.43 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr17_+_39737923 | 0.43 |
ENST00000577695.5
ENST00000309156.9 |
GRB7
|
growth factor receptor bound protein 7 |
| chr17_-_60392333 | 0.42 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr21_+_37367097 | 0.42 |
ENST00000644942.1
|
DYRK1A
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr16_-_72172135 | 0.41 |
ENST00000537465.5
ENST00000237353.15 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr9_+_27109135 | 0.40 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr2_-_164842011 | 0.40 |
ENST00000409184.8
ENST00000456693.5 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr2_-_65366650 | 0.40 |
ENST00000443619.6
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr2_-_74440484 | 0.40 |
ENST00000305557.9
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr17_-_42979993 | 0.39 |
ENST00000409103.5
ENST00000360221.8 ENST00000454303.1 ENST00000591916.7 ENST00000451885.3 |
PTGES3L-AARSD1
PTGES3L
|
PTGES3L-AARSD1 readthrough prostaglandin E synthase 3 like |
| chr7_-_151736304 | 0.39 |
ENST00000492843.6
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr4_+_168921555 | 0.39 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_42980393 | 0.38 |
ENST00000409446.8
ENST00000409399.6 ENST00000421990.7 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 like PTGES3L-AARSD1 readthrough |
| chr11_+_55811367 | 0.38 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr9_+_27109200 | 0.38 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr17_-_41055211 | 0.38 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr22_-_38302990 | 0.38 |
ENST00000612795.2
ENST00000451964.5 |
CSNK1E
|
casein kinase 1 epsilon |
| chr12_+_57583101 | 0.38 |
ENST00000674858.1
ENST00000675433.1 ENST00000674980.1 |
KIF5A
|
kinesin family member 5A |
| chr12_-_102120065 | 0.38 |
ENST00000552283.6
ENST00000551744.2 |
NUP37
|
nucleoporin 37 |
| chr10_-_77140757 | 0.37 |
ENST00000637862.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr7_+_66075876 | 0.37 |
ENST00000395332.8
|
ASL
|
argininosuccinate lyase |
| chr10_+_97572771 | 0.37 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr17_-_7179544 | 0.35 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chrX_-_30309387 | 0.35 |
ENST00000378970.5
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr14_-_58427134 | 0.35 |
ENST00000555930.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr18_-_63158208 | 0.35 |
ENST00000678301.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr11_+_122838492 | 0.35 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr14_-_80211472 | 0.35 |
ENST00000557125.1
ENST00000438257.9 ENST00000422005.7 |
DIO2
|
iodothyronine deiodinase 2 |
| chr2_+_172821575 | 0.35 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr19_-_2783241 | 0.35 |
ENST00000676943.1
ENST00000589251.5 ENST00000221566.7 ENST00000676984.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr2_-_69643152 | 0.34 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chr6_+_54111437 | 0.34 |
ENST00000502396.6
|
MLIP
|
muscular LMNA interacting protein |
| chr17_-_29761390 | 0.34 |
ENST00000324677.12
|
SSH2
|
slingshot protein phosphatase 2 |
| chr12_+_48482492 | 0.34 |
ENST00000548364.7
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr19_-_2783308 | 0.33 |
ENST00000677562.1
ENST00000677754.1 |
SGTA
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr11_+_57542641 | 0.33 |
ENST00000527972.5
ENST00000399154.3 |
SMTNL1
|
smoothelin like 1 |
| chr10_+_97572493 | 0.33 |
ENST00000307518.9
ENST00000298808.9 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr1_-_155910881 | 0.33 |
ENST00000609492.1
ENST00000368322.7 |
RIT1
|
Ras like without CAAX 1 |
| chr2_-_164842140 | 0.33 |
ENST00000496396.1
ENST00000629362.2 ENST00000445474.2 ENST00000483743.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr6_+_20401864 | 0.33 |
ENST00000346618.8
ENST00000613242.4 |
E2F3
|
E2F transcription factor 3 |
| chr20_+_1894145 | 0.33 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha |
| chr3_-_190449782 | 0.33 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr17_+_6996042 | 0.33 |
ENST00000251535.11
|
ALOX12
|
arachidonate 12-lipoxygenase, 12S type |
| chr8_-_130016395 | 0.32 |
ENST00000523509.5
|
CYRIB
|
CYFIP related Rac1 interactor B |
| chr14_-_58427158 | 0.32 |
ENST00000555097.1
ENST00000556367.6 ENST00000555404.5 |
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr3_+_141386393 | 0.32 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr14_-_58427114 | 0.31 |
ENST00000556007.6
|
TIMM9
|
translocase of inner mitochondrial membrane 9 |
| chr1_+_39081316 | 0.31 |
ENST00000484793.5
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr11_+_55827219 | 0.31 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr17_-_29761415 | 0.31 |
ENST00000649863.1
|
SSH2
|
slingshot protein phosphatase 2 |
| chr11_+_1838970 | 0.31 |
ENST00000381911.6
|
TNNI2
|
troponin I2, fast skeletal type |
| chr22_-_29388530 | 0.31 |
ENST00000357586.7
ENST00000432560.6 ENST00000405198.6 ENST00000317368.11 |
AP1B1
|
adaptor related protein complex 1 subunit beta 1 |
| chr13_+_75549734 | 0.31 |
ENST00000563635.5
ENST00000377595.8 |
ENSG00000261553.5
UCHL3
|
novel transcript ubiquitin C-terminal hydrolase L3 |
| chr10_+_73998104 | 0.30 |
ENST00000372755.7
ENST00000211998.10 |
VCL
|
vinculin |
| chr3_-_196287649 | 0.30 |
ENST00000441879.5
ENST00000292823.6 ENST00000431016.6 ENST00000411591.5 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr19_+_39498938 | 0.30 |
ENST00000356433.10
ENST00000596614.5 ENST00000205143.4 |
DLL3
|
delta like canonical Notch ligand 3 |
| chr6_-_88963409 | 0.30 |
ENST00000369475.7
ENST00000538899.2 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr5_+_40679907 | 0.30 |
ENST00000302472.4
|
PTGER4
|
prostaglandin E receptor 4 |
| chr10_+_63133279 | 0.30 |
ENST00000277746.11
|
NRBF2
|
nuclear receptor binding factor 2 |
| chr6_-_127519191 | 0.30 |
ENST00000525778.5
|
SOGA3
|
SOGA family member 3 |
| chr10_+_63133247 | 0.29 |
ENST00000435510.6
|
NRBF2
|
nuclear receptor binding factor 2 |
| chr8_+_27774530 | 0.29 |
ENST00000305188.13
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr2_+_55232672 | 0.29 |
ENST00000404735.1
ENST00000272317.11 |
RPS27A
|
ribosomal protein S27a |
| chr14_-_80211268 | 0.29 |
ENST00000556811.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr20_-_13990609 | 0.29 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr11_+_113908983 | 0.29 |
ENST00000537778.5
|
HTR3B
|
5-hydroxytryptamine receptor 3B |
| chr19_-_46471484 | 0.29 |
ENST00000313683.15
ENST00000602246.1 |
PNMA8A
|
PNMA family member 8A |
| chr6_-_88963573 | 0.28 |
ENST00000369485.9
|
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr2_+_196713117 | 0.28 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150 |
| chr19_-_46471407 | 0.28 |
ENST00000438932.2
|
PNMA8A
|
PNMA family member 8A |
| chr7_+_102748972 | 0.28 |
ENST00000413034.3
ENST00000409231.7 ENST00000418198.5 |
FAM185A
|
family with sequence similarity 185 member A |
| chr8_-_130016622 | 0.28 |
ENST00000518283.5
ENST00000519110.5 |
CYRIB
|
CYFIP related Rac1 interactor B |
| chr1_+_184051865 | 0.28 |
ENST00000644815.1
|
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chrX_+_71301742 | 0.28 |
ENST00000373829.8
ENST00000538820.1 |
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr1_+_15684284 | 0.28 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr9_-_120793377 | 0.28 |
ENST00000684001.1
ENST00000684405.1 ENST00000608872.6 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr5_-_163460048 | 0.27 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr1_+_20070156 | 0.27 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V |
| chr1_+_184051678 | 0.27 |
ENST00000643231.1
|
TSEN15
|
tRNA splicing endonuclease subunit 15 |
| chr22_-_19178402 | 0.26 |
ENST00000451283.5
|
SLC25A1
|
solute carrier family 25 member 1 |
| chr19_+_19528861 | 0.26 |
ENST00000436027.9
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr9_+_128552558 | 0.26 |
ENST00000372731.8
ENST00000630804.2 ENST00000372739.7 ENST00000627441.2 ENST00000358161.9 ENST00000636257.1 |
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr19_+_589873 | 0.25 |
ENST00000251287.3
|
HCN2
|
hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2 |
| chr15_-_79923647 | 0.25 |
ENST00000485386.1
ENST00000479961.1 ENST00000494999.1 |
ST20
ST20-MTHFS
|
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr3_-_194486945 | 0.25 |
ENST00000645538.1
ENST00000645319.2 |
ATP13A3
|
ATPase 13A3 |
| chr16_+_85908988 | 0.25 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr19_+_19528901 | 0.25 |
ENST00000514277.6
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr13_-_33185994 | 0.24 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr17_-_41065879 | 0.24 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr8_+_144095054 | 0.24 |
ENST00000318911.5
|
CYC1
|
cytochrome c1 |
| chr11_-_35360050 | 0.24 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_-_29181809 | 0.24 |
ENST00000466448.4
ENST00000373795.7 |
SRSF4
|
serine and arginine rich splicing factor 4 |
| chr9_-_120793343 | 0.24 |
ENST00000684047.1
|
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr19_-_48673017 | 0.24 |
ENST00000270235.11
ENST00000596844.5 |
NTN5
|
netrin 5 |
| chr7_+_45574358 | 0.23 |
ENST00000297323.12
|
ADCY1
|
adenylate cyclase 1 |
| chr2_-_229921903 | 0.23 |
ENST00000389045.7
ENST00000409677.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr2_+_74530018 | 0.23 |
ENST00000437202.1
|
HTRA2
|
HtrA serine peptidase 2 |
| chr16_-_68015911 | 0.23 |
ENST00000574912.1
|
DPEP2NB
|
DPEP2 neighbor |
| chr1_+_28438104 | 0.22 |
ENST00000633167.1
ENST00000373836.4 |
PHACTR4
|
phosphatase and actin regulator 4 |
| chrX_+_68693629 | 0.22 |
ENST00000374597.3
|
STARD8
|
StAR related lipid transfer domain containing 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.9 | 3.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.9 | 6.0 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.7 | 2.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.4 | 1.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.4 | 1.6 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 1.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 1.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.3 | 2.0 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.3 | 0.9 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 0.3 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.3 | 1.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 2.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.6 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.2 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 1.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.7 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.2 | 0.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 2.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.4 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 2.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 2.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.4 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.3 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.4 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 1.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 2.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 1.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 1.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0090133 | corticotropin hormone secreting cell differentiation(GO:0060128) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 1.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.5 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.2 | GO:0014016 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.8 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.5 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 2.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.4 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.6 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 1.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.4 | GO:0030238 | male sex determination(GO:0030238) Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 1.7 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 3.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 1.2 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 1.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 1.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.4 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.1 | 0.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 2.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 1.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 2.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 6.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.6 | 6.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.4 | 6.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 1.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 1.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 1.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 2.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.6 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.2 | 0.5 | GO:0044714 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.2 | 0.5 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.2 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 1.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0004056 | argininosuccinate lyase activity(GO:0004056) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 1.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 2.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.5 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 4.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 5.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 5.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 6.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |